eISSN: 2576-4462


Research Article Volume 2 Issue 5
Genetics and Agricultural Biotechnology Institute of Tabarestan, Sari Agricultural Sciences and Natural Resources University, Iran
Correspondence: Behzad Shahin Kaleybar, Genetics and Agricultural Biotechnology Institute of Tabarestan, Sari Agricultural Sciences and Natural Resources University, Iran, Tel 98 9141290839
Received: August 17, 2017 | Published: October 11, 2018
Citation: Kaleybar BS, Nematzadeh GA, Ghasemi Y, et al. Assessment of genetic diversity and fingerprinting of strawberry genotypes using inter simple sequence repeat marker. Horticult Int J. 2018;2(5):264-269. DOI: 10.15406/hij.2018.02.00062
In this study, the ability Inter Simple Sequence Repeat markers (ISSR) were used to identify molecular relationships among wild, local and commercial genotypes of strawberry. Twelve out of the 25 tested primers produced clear and reproducible fragments with high value of polymorphism (96.5%). The number of bands per primer ranged from 6 to 18 with an average of 13.16 bands. Clustering based on Jaccard's similarity coefficient divided the genotypes into five groups and separated wild accessions from others. The highest genetic distance was between Kaleybar and Pahnekola sari accessions with 0.31 similarity index that is promising to obtain hybrids with greater heterotic effect and segregation. The results indicated that there are high value of divergence between the genotypes and clearly demonstrate that ISSR markers can be used in a genetic diversity studying as well as in genotypic identification of strawberries. The results of the study can be use for further analysis, including fingerprinting, preservation of genetic resources, parental selection and hybrid/cross-breeding programs.
Keywords: fingerprinting, genetic diversity, genetic relatedness, issr marker, strawberry, wild species
ISSR, inter simple sequence repeat markers; RAPD, random amplified polymorphic dna; ROS, reactive oxygen species; SSR, simple sequence repeats or microsatellites; AFLP, amplified fragment length polymorphism; PIC, polymorphic information content; UPGMA, unweighted pair group method with arithmetic mean
Ability of a plant species to respond adaptively to environmental challenges depends on its genetic diversity.1 Strawberry is an economically and commercially important horticultural crop with rich source of bioactive compounds that are beneficial to human health.2 It has been reported that strawberry fruits, because of its high levels of vitamin C and K, folate, phenolic compounds and flavonoids, retards age-related effects on memory.3 Fruits of this horticultural crop are widely consumed fresh or in processed forms, such as jams, juices, and jellies. Strawberry fruits also has shown antioxidant and anti-cancer properties by inhibiting production of Reactive Oxygen Species (ROS) and carcinogens reduction.4–6 The diversity and high properties value of its compounds make strawberry a very attractive fruit for studying. Wild strawberry species as genetic resources are valued by breeders to produce new varieties with novel traits that are more productive, more nutritious, more market-friendly and more resistant to biotic (viruses, fungi, bacteria, weeds, insects and pests) and abiotic (drought, salinity, cold, heat) stresses. There are several systems such as morphological, chemical, and biochemical markers for evaluating diversity levels in plants. But these systems of classification are influenced by factors like temperature, humidity, light and/or plants ages which can modify results of classification. While, DNA-based marker systems provide a reliable and powerful tools for assessing differences between organisms with simultaneous elimination of the other systems constraints and are increasingly used in breeding programs and germplasm management of many horticultural crops. Several PCR (polymerase chain reaction)-based DNA marker systems including RAPD (random amplified polymorphic DNA), AFLP (amplified fragment length polymorphism) and SSRs (simple sequence repeats or microsatellites) are available for genetic assessment,7 but each of the methods have some limitations: low primer annealing temperature and reproducibility for RAPD, requirement for prior sequence information from flanking regions to develop primers for SSR and high experiment costs for AFLP. ISSR marker is a cheap, fast and simple genotyping technique based DNA that requires small amounts of DNA template.8 This marker is more reliable than RAPD because of longer length of primers and high annealing temperature and ISSR does not requires any prior sequence information. ISSR marker uses a single primer targeting microsatellite motifs that generates abundant polymorphic bands with a reliable and reproducible banding patterns in many systems.7,9,10 ISSR marker has been used successfully to assess genetic variation in a vast range of plants and horticultural crops including blueberry,11 lingo berry,12 citrus,13 potato,14 Oryza15 and described as a powerful technique to assess genetic diversity to detect similarities between and within species levels. It is well know that availability and deep knowledge of genetic diversity of any given crop will enhance extent of any improvements.16 Strawberry belongs to Rosaceae family, which has approximately 3000 members,17 Fragaria genera and six species all iedentifies as straswery. Strawberry has different size, color, taste, form, season of ripening, level of fertility and resistance to disease.18 One of the most important habitats for wild strawberries genotypes in Iran is northern strip, but there are little information on their genetic diversity and molecular relationships. The aim of this study was to evaluate the ISSR markers ability to differentiate strawberry genotypes, fingerprinting some Iranian varieties and determine their molecular relationships. This study is the first report to evaluate ISSR maker to determine the genetic diversity among wild north strip accessions and local varieties of Iranian strawberry genotypes and commercial cultivars. The results can be used to further studies like genetic diversity preservation, breeding programs and genetic or genomic studies.
Plant materials and genomic DNA isolation
In this study sixteen genotypes of strawberry including 9 wild accessions, 4 local varieties and 3 commercial cultivars were used to genetic relationships analysis. The local varieties were (Dalndi, Kordestan, Atabaki and Gilasi), commercial cultivars were (Camarosa, Diamante and Selva) and wild accessions collected from several region of Iran including pasand behshahr, Gorgan, pahnekola Sari, Kaleybar, Ramsar, Lahijan, hezarjarib, savadkoh and zirab (Figure 1). In order to DNA extraction, the seeds of the genotypes were removed from the fruits and cultured in greenhouse in a controlled condition (temperature, 29°C day and 27°C night; relative humidity, 60% day and 75% night; 16h illumination per day). After 50 days young leaves were collected from actively grown shoots, immediately frozen in liquid nitrogen and then placed at -80°C until DNA extraction. Genomic DNA was extracted using DNeasy Plant Mini Kit (QIAGEN, Germany) from approximately 0.1g of leaf material powdered by mortar and pestle according to manufacturer’s instructions. Quantity and quality of isolated DNA was determined by 0.7% agarose gel electrophoresis using known amounts of diluted uncut lambda DNA as standard and spectrophotometer analysis. The ratio of the absorbance at A260/A280 nm was used for measuring extracted DNA Purity and the samples with purity ratio between 1.8–2 were selected for subsequent applications and analyzing by ISSR marker.


Figure 1 Collection sites of strawberry accessions from northern strip of Iran. pasand behshahr (1), Gorgan (7), pahnekola Sari (5), Kaleybar (15), Ramsar (8), Lahijan (2), hezarjarib (12), savadkoh (13) and zirab (16).
ISSR amplification
Twenty-five ISSR primers were screened to determine their ability for detecting polymorphism among genotypes. Optimization of Polymerase chain reaction (PCR) amplifications were accomplished by changing concentrations of MgCl2, genomic DNA and annealing temperature. Finally the PCR reactions were performed in a volume of 20 µl reaction mixture containing 60ng of template DNA, 0.6mM of each dNTPs, 1XTaq buffer PCR (10mM Tris-HCl, 50mM KCl (pH 8.8) and 0.08Nonidet P40), 2mM MgCl2, 1U of Taq DNA polymerase and 0.4µM of the ISSR primer. To reduce background amplification, 2 % (v/v) formamide was added to the reaction mixture.19 Amplification reactions were carried out using ABI Applied Biosystems thermal cycler with an initial denaturation step of 5min at 95˚C, followed by 35 cycles; each cycle included 94°C for 40s, 55°C for 1min, 72°C for 2 min and a final extension step at 72°C for 7min. To test DNA contamination of the reagents a reaction, containing all PCR components except DNA (negative control) was used in each experiment. PCR amplified fragments were separated on 2.5% agarose gels by electrophoresis at 90V in 0.5xTBE (Tris / Boric Acid / EDTA) buffer. Gels were stained with ethidium bromide20 and imaged in Doc-Print VX5 (VIBER LOURMAT) gel documentation system.
Data analysis
ISSR reproducible fragments were scored as presence (1) or absence (0) for each of the possible band. Since ISSR is a dominant marker, the presence of a band was interpreted as either a heterozygote or dominant homozygote and the absence of a band as recessive homozygote. The data matrix structure was assembled by binomial (0/1) data and used as input data for further analysis using NTSYS version 2.02 software program.21 Cophenetic matrices were created to test whether clusters in the dendrogram agreed with information from the similarity matrix. Similarity for ISSR data was computed using the Jaccard’s similarity index and cluster diagrams were generated with the un-weighted pair group method using arithmetic averages (UPGMA) algorithm. The resulting clusters were expressed as dendrogram.
Initially to screen the best primers based on their ability to amplify DNA bands we used 25 ISSR primers that twelve out of which produced clear and reproducible fragments with multiband patterns and were considered as informative and polymorphic primers for further analysis that has been mentioned in Table 1. Within the selected primers there were ten di-nucleotide repeat and two tri-nucleotide repeat primers with 3’ anchors, remaining primers produced faint or no distinct bands on gel. For example the amplified bands by primer ISSR-2 has been shown in Figure 2. The primers used differed in size of amplified products between 190-2350 bp (Table 1) and no band was detected in any negative control amplifications. The primer #ISSR-19 exhibited the highest size variation of amplified products (300-2350bp) and the smallest one was observed for primer #ISSR-1 (190-840bp). Primer #ISSR-1 also produced the highest number of loci (18 loci) and the lowest one was amplified by primer ISSR-5 (6 loci) and the average bands per primer was 13.16. In total 155 loci were visualized with polymorphism range between 80 to 100 % with an average of 96.5 % that representing higher value of divergence between the genotypes and a high efficiency of the used primers to distinguish between the strawberry genotypes that the haracteristics of the primers have been shown in Table 2. Polymorphic Information Content (PIC) is a parameter that refers to value of a marker for detecting degree of polymorphism within a population and depends on number of detectable alleles and the distribution of their frequency.22 Analysis of the data showed that the PIC value varied from 0.18 to 0.31, with an average of 0.24. Primer #ISSR-13 by showing the highest PIC value (PIC=0.31) (Table 3) was recognized as appropriate and discriminating primer to estimate genetic similarity and diversity of the strawberry genotypes and used as a good primer for fingerprinting of the cltivars that its sequence has been presented in Figure 2.
|
Primer |
Sequence (5' to 3') |
Annealing temperature (°C) |
Fragment size length |
Total no. of bands |
No. of Polymorphic bands |
Polymorphism (%) |
PIC |
|
ISSR-1 |
(GA)9A |
55 |
190-840 |
18 |
18 |
100 |
0.23 |
|
ISSR-2 |
(GA)9C |
57 |
250-1500 |
11 |
11 |
100 |
0.27 |
|
ISSR-5 |
(GT)9C |
57 |
470-1300 |
6 |
6 |
100 |
0.28 |
|
ISSR-8 |
(CT)8G |
51 |
320-1620 |
14 |
14 |
100 |
0.27 |
|
ISSR-9 |
(AG)8C |
51 |
265-1200 |
15 |
14 |
93 |
0.19 |
|
ISSR-11 |
(GA)8C |
51 |
300-1500 |
16 |
16 |
100 |
0.24 |
|
ISSR-13 |
(TC)8C |
51 |
570-1670 |
8 |
7 |
80 |
0.31 |
|
ISSR-14 |
(TC)8G |
51 |
560-1580 |
14 |
14 |
100 |
0.18 |
|
ISSR-16 |
(TG)8A |
50 |
450-1840 |
13 |
13 |
100 |
0.21 |
|
ISSR-17 |
(AC)8C |
51 |
485-1820 |
15 |
15 |
100 |
0.2 |
|
ISSR-18 |
(ATC)6T |
49 |
200-1450 |
14 |
14 |
100 |
0.24 |
|
ISSR-19 |
(ATC)6C |
51 |
300-2350 |
14 |
12 |
85 |
0.3 |
Table 1 The list and features of ISSR primers used in this
|
Dice’s similarity |
Jaccard’s similarity |
SM’s similarity |
UPGMA |
0.86895 |
0.8858 |
0.67998 |
UPGMC |
0.86895 |
0.53729 |
0.29065 |
COMPLETE |
0.80569 |
0.83188 |
0.66416 |
SINGLE |
0.84211 |
0.85581 |
0.6909 |
Table 2 Calculated Cophenetic correlation for each method combination
Cultivers |
ISSR primers(bp) |
||||||||||||
ISSR-2 |
ISSR-9 |
ISSR-13 |
ISSR-16 |
ISSR-18 |
|||||||||
1000bp |
1150 bp |
400 bp |
500 bp |
820 bp |
410 bp |
1000 bp |
1350 bp |
700 bp |
950 bp |
250 bp |
1200 bp |
800 bp |
|
Dalandi |
– |
⁺ |
– |
– |
– |
– |
– |
⁺ |
⁺ |
⁺ |
⁺ |
– |
– |
Kordestan |
– |
– |
– |
⁺ |
– |
⁺ |
⁺ |
– |
– |
– |
⁺ |
– |
– |
Atabaki |
⁺ |
⁺ |
⁺ |
⁺ |
⁺ |
⁺ |
– |
⁺ |
– |
– |
– |
– |
⁺ |
Gilasi |
– |
⁺ |
– |
– |
– |
⁺ |
⁺ |
– |
⁺ |
– |
– |
⁺ |
⁺ |
Table 3 Identification of four cultivars on the basis of the presence (+) or absence (-) of ISSR markers
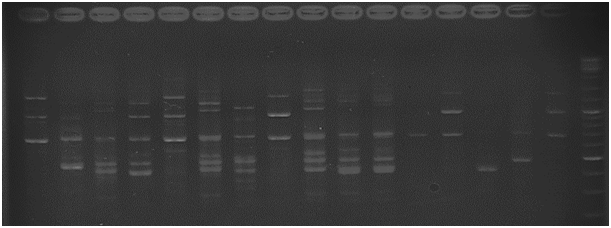
Figure 2 The amplification profile of strawberry genotypes using primer 2 and separated in 2.5% agarose gel.
Genetic relationships
The diversity data obtained from similarity analysis were varied between 0.31 to 0.70 with an average of 0.43 The high genetic divergence found may have been due to a highly heterozygote genetic structure among the strawberry genotypes because of their different origins and ploidy levels. The highest similarity index was between Camarosa (10) and Diamant (11) cultivars with value of 70 %. These cultivars came from the same breeding programs and were developed at the University of California and may share ancestors. Morales et al.23 in studying of genetic diversity among strawberry cultivars using ISSR and RAPD markers, have reported high similarity index between Camarosa and Diamant cultivars with value of 67% similarity. The lowest similarity index was between pahnekola sari (5) and Kaleybar (15) accessions with value of 31% similarity. This degree of dissimilarity may be cause of a diverse genetic background of Kaleybar accession because of its geographical distance (Figure 3). In plant breeding programs the genotypes that have the highest genetic distance and high means of performance for the target traits are used as parents to crosses with adapted varieties for obtaining hybrids with greater heterotic effect and segregation.24–26 In this sense, it could be inferred that crosses involving the Kaleybar genotype because of its good characteristics such as high resistance to cold stress and lowest similarity (60%) to the other genotypes is the most interesting.

Figure 3 Constructed dendrogram based on Jaccard’s similarity coefficient and UPGMA algorithm to show the genetic relationships among 16 genotypes of Strawberry by ISSR marker.
Cluster analysis
In order to distinguish the best methods for clustering and similarity coefficient calculation, the cophenetic correlation, a measure of the correlation between the similarity represented on the dendrogram and the actual degree of similarity, was calculated for each method combination (Table 2). The UPGMA clustering method based on Jaccard’s similarity coefficient, exhibited the highest value of cophenetic correlation (r=0.88), Therefore the dendrogram constructed based on this method was used for representing genetic diversity of genotypes (Figure 4). Multivariate statistics such as cluster and principal components analysis play a principle role in investigation genetic distance diversity and evolution of plants. Assessment of Genetic diversity is important not only for efficient management and conservation of germplasm resources but also is essential for crop improvement. Cluster analysis based on the genetic similarity divided the strawberry genotypes into five groups at the cutoff level of 43% similarity. Group I contains 6 accessions out of 9 wild accession (Pasand behshahr, Pahnehkola sari, Ramsar, Hezarjarib, Savadkoh, Zirab) that indicates the ability of the ISSR marker for strawberry genotypes genetic diversity studying. Group II contains all commercial genotypes (i.e. Camarosa, Diamante and Selva) and local varieties (Dalandi, Kordestan, Atabaki and Gilasi). It should be noted that there are no detailed reports on genetic backgrounds and origins of local varieties that are grown in Iran. The dendrogram and similarity data analysis exhibited a close genetic background of Iranian local varieties with commercial cultivars especially with Camarosa. These results raise the possibility of a common ancestor among them. Groups III, IV and V each contain only one wild accession that was coherent with their origins. Similarity data analysis showed a high level of genetic distances among the three Iran wild accessions of northern strip (Groups III, IV and V) with other ones that is indicative of their different genetic backgrounds. It may be due to geographical distance of the genotypes because they were collected from different provinces. To show multiple dimensions distribution of the genotypes and better explaining genetic variation in a scatter-plot, principal coordinate analysis was performed on the basis of Jaccard similarities. The first, second and third components represented 12%, 10% and 9% of the total variance respectively and the first 10 coordinates explained 78% of total variation that are indicative of appropriate sampling of primers and scattering them over different parts of the genome.
None.
Authors declare that there is no conflict of interest.

©2018 Kaleybar, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.