eISSN: 2469-2794


Research Article Volume 7 Issue 3
Beijing Institute of Radiation Medicine, China
Correspondence: Xiao Lei, Beijing Institute of Radiation Medicine, Beijing, P.R. China
Received: May 08, 2019 | Published: June 19, 2019
Citation: Lei X. Comparison of different STR typing for a special allele at D7S820 locus by using ten different STR multiplex system. Forensic Res Criminol Int J .2019;7(3):135-142. DOI: 10.15406/frcij.2019.07.00277
During a DNA database construction and population study in China, we observed a special genotype 10/10.1 at D7S820 locus which was misjudged by seven out of ten different STR multiplex systems. Allele typing results acquired by using ten diverse STR multiplex kits (PowerPlex® Fushion6C, PowerPlex® 21, PowerPlex® 24, ACGU® EX25, AppliedBio® HuaXiaBaijin, SureID® PanGlobal, GoldeneyeTM 20A, GoldenEyeTM 25A, AppliedBio® GlobalFiler, HuaDa® YanHuagn) and then analyzed by GeneMapper®1.4 software. In this paper, we conclude the reasons for the misjudgment of this allele as well as how to reduce these kinds of errors during STR analysis.
Keywords: short tandem repeat, D7S820, float bin, virtual bin, off-ladder, marker range, bin range, OL peak
Nowadays, more and more rare alleles are studied and supplemented to common Loci system, especially for the construction of DNA database and population studies. The ability to accurately judge the rare Loci becomes more essential. D7S820 is one of the useful markers for human identification, paternity and maternity testing and sex determination in forensic sciences.1 It has been revealed 4 microvariant alleles: 8.1, 9.1, 10.1 and 10.3.2 Here we designed a study to evaluate the performance of allele typing at D7S820 locus, with ten different STR multiplex systems, which mostly have been used in current forensic application for a long time. In this paper, we observed an abnormal genotype shown different results when tested by ten different STR multiplex kits.
Genetic characterization of one special individual sample was carried out using blood (DNA extraction: CN-QIAamp-DNA-Investigator) (Qiagen). PCR products of D7S820 were generated using following kits: PowerPlex® Fushion6C, PowerPlex® 21, PowerPlex® 24, ACGU®EX25, AppliedBio® HuaXiaBaijin, SureID® PanGlobal, GoldeneyeTM 20A, GoldenEyeTM 25A, AppliedBio® GlobalFiler, HuaDa® YanHuagn. Automated fragment analysis was carried out on the ABI 3500 Genetic Analyzers (Applied Biosystems, Foster City, CA, USA). Allele designations were automatically assigned by GeneMapper 1.4 Software and every kit above by size comparison between sample alleles and allelic ladder alleles, run on the same gel or set of injections. Peaks are labled with the allele category and the calculated fragment size using the internal sizing standard (Liz600) with GeneMapper 1.4. The direct Taq-cyclesequencing method was performed.3‒7
Parallel tests with AppliedBio®GlobalFiler shows two different allele typing: homozygote-10/10, 10.1/10.1 and likely split peaks. So we performed re-examination by using PowerPlex systems (PowerPlex® Fushion6c, PowerPlex® 21, PowerPlex® 24) (Figure 1 & Figure 2).
PowerPlex® 21 (heterozygote 9.3/10.1) (lower left); PowerPlex® 24 (heterozygote 10/10.1) (lower right) show different results. Same sample was tested by all three kinds of PowerPlex kits: PowerPlex® Fushion6C, PowerPlex® 21 and PowerPlex® 24. Although, clearly, there is a split peak at this locus, two different heterozygous typing are observed: the first two kits have identical results:9.3/10.1. and the other kit is 10/10.1. Then we applied six other multiplex kits including ACGU® EX25, AppliedBio®HuaXiaBaijin, SureID® PanGlobal, GoldenEyeTM 20a, GoldenEyeTM25a and HuaDa ® YanHuagn to find out the real peak-shape and the right typing of this rare allele (Figure 3).
After several repeated experiments were settled, the possibility of contamination was eliminated. From Figure 3 we can see that results of six kits are also varied from both peak shape and allele typing. Single-peak and double-peak pattern were observed and the typing include both homozygote and heterozygote. Some of the heterozygote typing have one OL (off ladder) peak within the white longitudinal stripe Figure 3(A‒D), which means they are out of marker/bin range and cannot be marked by the system ladder automatically. Although we can label the off-ladder allele 10.1 manually, we still leave them unmarked Figure 3(A‒D) for parallel comparison. In order to find out the real typing of this rare allele, we also applied Sanger Sequencing test.8,9
10.1:GGGGCTAACGCAGTGCAGCTTGCATGCCTGCAGGTCGACGATTGACCCCCTATGGAATTTTTTTGTTTGTTTGTTTTTATTTATTTCTTTATCTTGAGATGGAGTCTCACTCTGTCACCCAGGCTGGAGTGCAGTGGTGCGATCTCGGCTCACTGCAACCTCCGCTTCTTGGGTCAAGTGGTTCTCCTGCCCCAGCCTCCTGAGTAGCTGGGACTACAGGCATGTGCTACTGCATCCAGCTAATTTTTGTATTTTTTTTAGAGACGGGGTTTCACCATGTTGGTCAGGCTGACTATGGAGTTATTTTAAGGTTAATATATATAAAGGGTATGATAGAACACTTGTCATAGTTTAGAACGAACTAACA(GATA)10GACAGATTGATAG(T)9ATCTCACTAAATAGTCTATAGTAAACATTTAATTACCAATATGTGGTGCAATTCTGTCAATGAGGATAAATGTGGAATCGTTATAATTCTTAAAAATATATATTCCCTCTGAGTTTTTGATACCTCAGATTTTAAGACCTCACAATTATCTCACAAGGCTTAAAATCAATCATATTTTGAGGATCACCTTATGGTATTTTTGCCTGTTTTTATTCCTTCTGGTGTGAAAACTGATGCCTTCCATCGTGTAACTCTTGTTCACACTGGTTTCAGTATTTTGTTTTGAATCTCTAGAGGATCCCCGGGTACCGAGCTCGAATTCGTAATCATGGTCATAGCTGTTTCCTGTGTGAAATTGTTATCCGCTCACAATTCCACACAACATACGAGCCGGAAGCATAAAGTGTAAAGCCTGGGGTGCCTAATGAGTGAGCTAACTCACATTAATTGCGTTGCGCTCACTGCCCGCTTTCCAGTCGGGAAACCTGTCGTGCCAGCTGCATTAATGAATCGGCCAACGCGCGGGGAGAGGCGGTTTGCGTATTGGGCGCTCTTCCGCTTCCTCGCTCAC
As we known, the rare allele 10.1 have two microvariations: TAACA (GATA)10 GACAGATTGATAG (T)9 and TAAC-(GATA)10GACAGATTGATAG(T)10.2 In our case, the sequence of this rare allele is TAACA (GATA)10 GACAGATTGATAG(T)9 (Figure 4), which is in concordance with the data reported by Tsuji et al.10 Hence, In previous tests of ten STR multiplex kits, only three kits achieved the correct typing (Here we only present sequencing result for the rare allele 10.1, no allele 10, since allele 10 was for sure in this locus), they are: HealthGene PanGlobal, HuaDa® YanHuagn, and AppliedBio® HuaXiaBaijin. (Although ACGU EX25 and GoldeneyeTM 20A have identical split peaks, one of the peak is still marked ‘OL’ and cannot be labeled by GeneMapper software, hence, they cannot be counted for correct results).
Generally speaking, STR multiplex system ladders were divided into three colored longitudinal stripe: Grey, Pink, and White. Grey means float bin, Pink for virtual bin, and White stripe is off-ladder area or out of marker range Figure 5.
Sample peaks within Grey or Pink stripes can be automatically marked by STR analysis software. In our tests, two ‘OL’ peaks fall outside of the bin range, means they are out of marker/bin range. For PowerPlex® Fushion6C and PowerPlex® 21), both ladders have a little deviation to the left, hence, their sample peaks are all near stripe border between virtual bin and off-ladder area which tend to be another source of error and could easily misjudged by GeneMapper software. Sample peaks can be marked within a bin range (ether float bin or virtual bin), but when outside these bins, they cannot be marked by GeneMapper or other STR analysis software automatically. However, an off-ladder peak can be marked manually by comparing the position of the peak on the panel with nearest bin on the ladder. Virtual bin centers were created using the offset value from a neighboring allele and the reference (sequence length) size of the virtual allele. For example, some virtual alleles either size within 4bps of the smallest or largest allelic ladder allele, or contain 2bps partial repeat units. Virtual bins for alleles containing 1bp or 3bps partial repeat units were not included. Integer designations for these variant alleles must be assigned manually. In addition to the substantial expansion of nearly 300 configured markers, support for novel microvariants has been included for all loci with expanded ‘virtual bin sets’ comprising each potential base call within the allelic range rather than only observed nominal allele bins.11
For some kits like ACGU EX25 and GoldeneyeTM 20A, their virtual bins for rare alleles like 10.1, are reduced or removed fromD7F820 locus, as for some rare allele no longer in bin/marker range and cannot be recognized by GeneMapper® or other STR analysis software automatically.
Manually analysis of an off-ladder allele at D7S820 locus with AGCU EX25 (Figure 7) is shown in GeneMapper Software plots (bp size versus relative fluorescence units). To do so, first we should know the repeat number of the certain locus. Let’s take the system panel of AGCU EX25 For example, the repeat base value of D7S820 labeled as ‘marker’ is ‘4’, which can be found in the upper middle ‘panel manager’ tab, marked by blue bar (Figure 6). This value also means microvarient alleles can only include 10.1,10.2 and 10.3. We can also check the label ‘Ladder Alleles’ to the right on the same blue bar of he tab. Allele 10.1 is not in the list, which means we must label this allele manually.
Sample allele was identified by recognition within floating bins (vertical gray stripes) around D7S820 alleles of the AGCU EX25 allelic Ladder Figure 7. One of the alleles that fell outside allelic ladder bins was flagged as ‘OL’ allele is shown. In this example, peak position is compared to the nearest allelic ladder using GeneMapper software. To be specific, as shown in Figures 7A & B, we put the mouse at off-ladder peak ‘OL’ and find the value of the ‘Size’ label in the lower left of the tab shows: 227.25bps. Then we put the mouse at the nearest ladder peak ‘10’, the value is 226.33bps.The apparent size difference relative to the D7S820 allele 10, verified by DNA sequencing as a 1bp insertion, defined a D7S820 10.1 allele. Sometimes, easier way is to use the abscissa value in lower left corner Figure 7C of the tab when pointing at the bin’s peak near the off-ladder peak instead of the ‘Size’ value of the ladder.
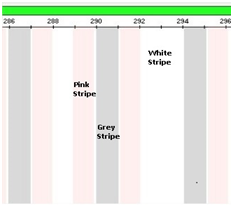
Figure 5 Grey (float bin), Pink (virtual bin), and White (no bin, out of marker range) longitudinal stripe on system marker.
Sometimes, off-ladder allele may fall out of the locus range on system panel Figure 8. Under these circumstances, we should judge which locus the off-ladder allele should belong to at first, by comparing the distances to each locus respectively. Normally, the closer the peak to the certain locus, the more chance it should belong to. Then we can compare the position of the off-ladder peak with the nearest bin with the method described above. In this case, the difference value is 8bps, which means there are 2 repeats between the off-ladder peak and the bin, thus, the off-ladder allele should be labeled as ‘8’. But the most reliable way to get this kind of off-ladder allele typing is to perform ether Sanger or massive parallel sequencing test.
However, even within bin range, results could be inaccurate (Figure 2). This mainly due to the peak deviation. Sometimes, ladders could have little deviation to the left or right from bins, hence, sample peaks from same batch could also have displacement in the same direction. When these peaks near stripe border between bins (gray or pink stripe) and off-ladder area (white stripe), they could be misjudged by GeneMapper software, error occurred. Peak deviation caused by many reasons, apart from experimental errors, one of the most important reasons is that the primer and position design of the locus on the panel was not optimized. Optimization for rare allele can either by adding virtual bins to the flank regions of neighboring loci or put loci with rare alleles within ranging of 75-250 bps in the system panel when designing multiplex primers.11 As we known, the best position for detecting rare alleles is within 75-250bps on the STR multiplex kit’s system panel. This is because the further away from this range, the speed of electrophoresis is getting more unstable, the peaks became wider and lower, and the system perform lesser resolution, especially for rare alleles, therefore, peak deviation appears.
Table 1 shows that except for AppliedBio® HuaXiaBaijin, two of the three kits* which achieved the correct typing are within preferred range of 75-250bps. Since AppliedBio® HuaXiaBaijin is specially designed for Chinese population, although it is not in the optimized range, there are certain virtual bins in locus with rare alleles like D7S820 which also contribute to the detection of rear alleles. Other two kits**, ACGU EX25 and GoldeneyeTM 20A, although have split-peak shape and within the preferred range, the 10.1 allele didn’t marked by GeneMapper software in short of certain virtual bins. But we can still achieve the correct typing by manual analysis as mentioned above. GoldeneyeTM25A only have a single peak, not even a trend of split-peak as GlobalFiler does. The main reason is because the position for locus D7S820 on the panel is around 350bps, far beyond the preferred detection range of 75-250bps. However, the genotype 10, 10.1 is tricky since if panel and locus bins for D7S820 are not optimized, presence of two alleles can be mistaken for inefficient non-template addition and split peaks. It is similar situation as in genotype with 9.3/10 allele of TH01 locus. Fortunately, we also detected this genotype with SureID®PanGlobal (Figure 9).
During STR test, when encountered with certain rare alleles, peaks could be easily misjudged as OLs (off ladders), since GeneMapper software cannot find corresponding data from the system ladder. To reduce these kinds of errors, companies should make necessary optimization including both putting loci with rare alleles into 75-250bps on the panel and adding certain virtual bins for rare alleles at the same time when designing STR multiplex kits. For certain population group, virtual bins for some specific loci should also be taken into account. Traditional capillary electrophoresis (CE) is still widely used for forensic DNA typing, mainly due to its time- and cost-effectiveness. Nowadays, native Bio-companies in China tend to add more and more numerous loci into 5- or 6-color fluorescence STR multiplex systems. On one hand, this may statistically increase the whole system accuracy. But on other hand, more rare alleles may appear and need to be optimized. The lack of optimization for rare alleles may inevitably cause more off-ladder peaks. Technical speaking, In order to avoid wrong judgement for rare alleles, kit designers should expand the assigned length on the panel and increase the number of virtual bins for every locus with rare allele in the system to cover all necessary known rare alleles (or rare alleles of certain population) to make those off-ladder rare allele peaks back into marker/bin range. For analyzers, the best way to avoid the error is manually analyze the typing of an off-ladder rare allele with the methods we mentioned above. An experienced lab staff could easily tell the real typing of an off-ladder rare allele by comprehensive analysis of different data like allele fragment length and peak position/shape via STR analysis software or by Sanger/Massive parallel sequencing test. In DNA database construction, rare allele types can greatly increase the power of discrimination. However, particular care should be taken in kinship matching and forensic cases, since incorrect designation of any deviations from allelic ladders could lead to a false conclusion.2 Therefore, it is necessary to increase the number of useful references on non-standard allele patterns.2 At the same time, parallel comparison is necessary to choose a suitable brand of STR multiplex kit as well as one or two standby kits for rare allele test during DNA database construction.
|
Multiplex systems |
D7F820 locus position on panel (bps) |
|
AppliedBio®GlobalFiler |
279 |
|
Promega PowerPlex®Fushion 6c |
290 |
|
PowerPlex®21 |
290 |
|
PowerPlex®24 |
290 |
|
ACGU EX25** |
227 |
|
AppliedBio®HuaXiaBaijin* |
279 |
|
SureID®PanGlobal* |
240 |
|
GoldeneyeTM20A |
226 |
|
GoldeneyeTM25A** |
351 |
|
HuaDa ®YanHuang* |
230 |
Table 1 Position designed for D7F820 locus on system panels of 10 different kits.
None
The author declares that there are no conflicts of interest.

©2019 Lei. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.