eISSN: 2473-0815


Research Article Volume 6 Issue 6
Instituto de Ensino e Pesquisa, Santa Casa de Misericordia de Belo Horizonte, Brazil
Correspondence: Karla Simone Fernandes, Instituto de Ensino e Pesquisa, Santa Casa de Belo Horizonte, P O, Box, 30250 140, Belo Horizonte, Brazil, Tel +55 31 981047557
Received: October 23, 2018 | Published: November 23, 2018
Citation: Rubatino FVM, Ferreira LA, Freitas ML, et al. Alterations in transcript levels of hypothalamic endosomal recycling genes in high-fat diet-fed mice. Endocrinol Metab Int J. 2018;6(6):429-432 . DOI: 10.15406/emij.2018.06.00229
It has been well established that body weight gain observed in the high-fat diet (HFD) murine models is associated with inflammation-induced disturbances in protein degradation, leading to neuronal damage. We hypothesized that HFD dysregulates the protein recycling machinery in the mice hypothalamus. Therefore, we determined transcript levels of endosomal sorting proteins in HFD C57BL/6J fed mice. A qPCR-array of retromer-wash and cargo protein genes was performed in the arcuate nucleus (ARC) isolated from mice that had received high-fat or standard diets (StD). We found that transcript levels of Ube2o, Tbc1d5, Igfr2, and Cdm6pr were negatively associated with weight gain and hyperglycemia. We concluded that HFD-induced weight gain altered transcript levels of endosome retrieval system genes, suggesting that it could be dysregulating the trafficking of steady-state surface receptors in the hypothalamus.
Keywords: obese mice, high-fat diet, hypothalamus, retromer-wash complex, satiety
The central nervous system (CNS) plays an important role in food intake and energy expenditure regulation.1 In the CNS, the ARC is suggested to be a key hub for energy balance, reducing food intake and improving energy expenditure through leptin and insulin interactions with their receptors, ObR and IR, respectively (Coll et al., 2007). An important step for physiological ligand-mediated cell signaling is the maintenance of a steady state of functional receptors at the plasma membrane, which involves dynamic intracellular transport mechanisms.2 Leptin and insulin receptors require internalization and retrograde routes upon the binding of their ligands,3 then sorted and transported by vesicles, for direct recycling to the plasma membrane or via the Trans-Golgi Network (TGN) or lysosomal degradation.4 Retromer, a protein trimer, is essential for the recruitment of nucleation-promoting factors to endosomes, including the WASH complex, which has a carboxy-terminal VCA motif that binds to actin, allowing subsequent actin filament nucleation by the Arp2/3 complex.5,6 On the other hand, non-recycled cargoes are directed to lysosomal degradation pathways. Newly synthesized hydrolases acquire a mannose-6-phosphate marker during their maturation and are then transported by the mannose-6-phosphate receptors (CDM6PR, CIM6PR also named IGFR2) in a step for lysosomal formation.7,8 In this study, we evaluated the transcript levels of genes involved in endosomal recycling in a murine model of HFD induced obesity and showed that HFD fed mice presented altered transcript levels for Ube2o, Igfr2, Cdm6pr and Tbc1d5. And showed significant body weight gain and hyperglycemia.
Experimental murine model
All animal procedures were approved by the ethics committee at IEPSCBH, 0001-15. StD (3.7kcal/g, 10% from saturated fat) and HFD (5.3 kcal/g, 60% from saturated fat), were purchased from PragSoluções Biociências, Brazil. Male, 4-week-old C57BL/6JUnib, had housing occupation up to four animals per cage. Temperature (22 ± 2°C) and light cycle (12-h light/dark) were controlled and animals had food and water ad libitum. Animals were randomized into two groups of six animals: one received 8 weeks of the HFD and the other receiving the StD for 8 weeks.
Transcript level analyses
Brains were removed from StD (n=6) and HFD (n=6) mice and immediately immersed in liquid nitrogen to preserve RNA. Hypothalamus were stored in RNAlater solution. RNA extraction was performed using the RNeasy Tissue Microarray (Qiagen®, Hilden, Germany), following the manufacturer's protocol. We performed transcript quantifications using a quantitative RT-PCR (qRT-PCR) assay with a two-step reaction process. cDNA synthesis was performed using the RT2 First Strand Kit (Qiagen®, Hilden, Germany). qPCR for the target genes was performed using a mouse customized RT2 Profiler PCR Array platform (Qiagen®, Hilden, Germany). Gene transcripts were determined according to Bustin et al.9
Statistical analysis
IBM SPSS Statistics 20 and R-3.3.0 software were used. A normal distribution was assumed for continuous variables followed by the Kolmogorov-Smirnov test. All data are represented as means ± standard deviations from a representative experiment of at least three independent experiments. Significance was accepted at p<0.05. For univariate analysis Student t-test was used. Bivariate comparisons were evaluated using the Pearson coefficient determination. For multivariate analysis, a Euclidean distance matrix was created. Ward’s method for hierarchical clustering was applied and the number of clusters was obtained considering the D index values.10
High-fat diet (HFD) induces weight gain and hyperglycemia in mice.
Mice receiving the HFD up to the 16th week were significantly heavier (42.3±4.3g; 45,8%) than mice that received the StD (29±1.9g; p<0.001; Figure 1A). They also presented significant hyperglycemia (192.3±27.3 mg/dL), when compared with StD mice (45.4±14.4 mg/dL; p<0.001; Figure 1B).
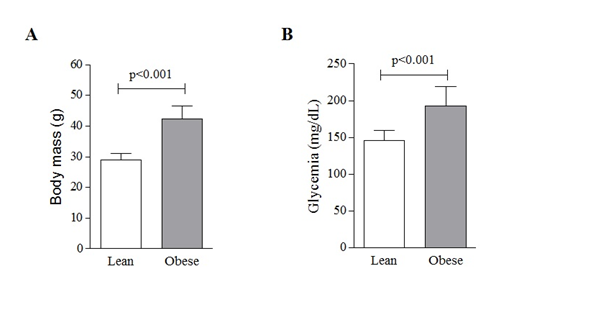
Figure 1 High-fat diet-induced obesity in mice. Twelve-week-old C57BL/6Junib male mice were randomly assigned to standard (St) (n=6) or high-fat diet (n=6). Body mass (A) and plasma glycemia (B) were determined on the 8th week. Values are means ± SD. t Student test was applied to compare means between groups. A representative of three independent assays.3.2. Transcript levels of endosomal protein sorting machinery are altered in HFD obese mice.
Mage-L2, Trim27, Wash, Fam21, Vps35, Kiaa1033 and Arpc2 genes presented unaltered transcript levels after HFD feeding (p>0.05; data not shown). Decreased transcript levels of Ube2o (62.3%; p=0.027), Igfr2 (64.1%; p=0.026), Cdm6pr (54.8%; p=0.002) and Tbc1d5 (67.1%; p=0.04) were observed in the hypothalamus of HFD mice, when compared to StD mice (Figure 2A). Total body weight negatively correlated to Ube2o (p=0.027), Igfr2 (p=0.038), Cdm6pr (p=0.003) and Tbc1d5 (p=0.03) levels, according to Pearson’s coefficient. Plasma glucose levels also negatively correlated to Ube2o (p=0.047), Igfr2 (p=0.026), Cdm6pr (p=0.008) and Vps35 (p=0.01) levels.

Figure 2 Determination of Ube2o, Igfr2, Cdm6pr and Tbc1d5 mRNA levels in the arcuate nucleus by qPCR. Twelve-week-old C57BL/6Junib male mice were randomly assigned to a standard (St) (n=6) or high-fat diet (n=6). Gene expression data were determined by qPCR on the 8th week and represented as fold change relative quantitation. Values were expressed in means ± SD. Comparisons between groups were evaluated by one-way analysis of variance, followed by the Student T-test. p<0.05 was considered significant. For both groups, n=6.
High-fat diet determines phenotypes by cluster definition based on Ube2o, Tbc1d5, Igfr2 and Cdm6pr transcript levels in the hypothalamus
We performed a multivariate analysis to determine whether the HFD was able to induce alterations in gene expression profiles in the hypothalamus. This approach provides a graphical visualization of intragroup homogeneity and intergroup heterogeneity10 and this clustering method defined three distinct phenotypes for the Ube2o, Tbc1d5, Igfr2 and Cdm6pr transcript levels, considering the D index (Figure 3A).
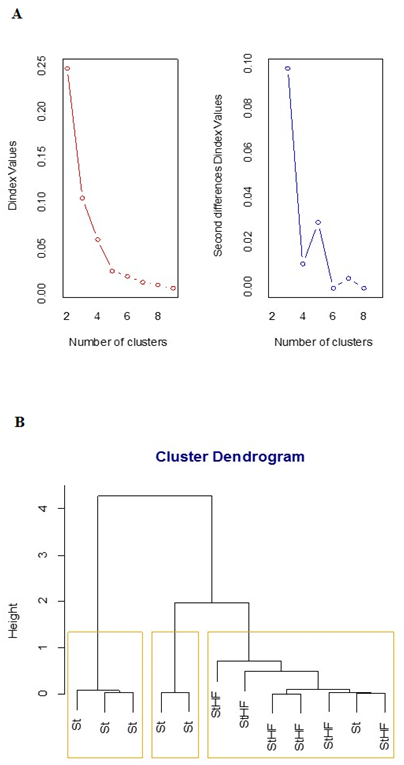
Figure 3 Hierarchical Clustering for St and StHF feed mice by Ward’s method. Twelve-week-old C57BL/6Junib male mice were randomly assigned to a standard (St) (n=6) or high-fat diet (StHF; n=6). Fold change values were determined by qPCR on the 8th week. Euclidean distance was used as metric with UBE2O, IGFR2, CDM6PR and TBC1D5 fold change values as dimensions (clustering dendrogram). The number of clusters was obtained by D Index.
This is the first study reporting retromer-wash complex regulation in HFD fed mice. Our main finding is that body weight gain induced by HFD alters transcript levels of genes involved in the endosome-to-Golgi retrieval pathway. More specifically, transcript levels of the cargo proteins, Cdm6pr and Igfr2, Rab-GAP Tbc1d5 and Ube2o E2-conjugase, were significantly decreased in HFD obese mice. The endosome-to-Golgi retrieval pathway is suggested to form part of the recycling machinery of a variety of proteins, from early to late endosomes and back to TGN, the plasma membrane and lysosomal degradation, and is essential for diverse cellular functions.5,11,12 Indeed, we specifically targeted some genes involved in endosomal protein sorting. They have been also associated with neuronal functions13 and neurodegenerative disorders.11,12 Quantitative PCR analyses showed that Ube2o, Igfr2 and Tbc1d5 Rab-GAP are decreased in the hypothalamus of obese mice and are inversely correlated with glycemia. Ube2o and Mage-L2-Trim27 siRNA knockdown has been shown to impair retromer complex activation,5 where both Mage-L2-Trim27 and Ube2o are essential for Igfr2 (cargo) endosome-to-Golgi retrograde transport. We found no alteration in hypothalamic Mage-L2 and Trim27 levels in HFD fed mice, and no differences in WASH components.
Our findings concerning altered Tbc1d5 levels suggest a dysfunction in retromer complex recruitment. It is known that retromer recruitment for the sorting of Igfr2 to lysosomes is mediated by Rab7 and inhibited by the GTPase effect of Tbc1d5.14 In addition, the disruption of one step in retrograde transport is thought to be sufficient to impair all recycling of cargo proteins.5
It is well established that obesity induced by high-fat diet is accompanied by insulin and leptin resistance in mice15-19. The limitations of our study were the lack of leptin and insulin resistance measurements or fat mass and food intake indexes. We chose body weight gain as a parameter for associating with endosome trafficking gene expression and found increases in mean body total weight gain and hyperglycemia that were consistent with those of previous reports.15,19,20 Thus, although our study did not investigate ObR expression in the hypothalamus of obese mice, reduced expression of Igfr2 in HFD fed mice may suggest some alteration in ObR retrograde transport to TGN. ObR is reported to be endocytosed in a ligand-independent manner and colocalizes mainly with IGFR2 in the TGN compartment.1 Furthermore, ObR is postulated to be retained intracellularly, suffering subsequent lysosomal degradation, which could maintain ObR surface expression at low levels in an in vitro and controlled milieu.21 The dynamics of ObR activity have also been elucidated in diet-induced obesity (DIO) mice, where both ObR cell surface density and STAT3 signaling activity were diminished in HFD mice.21,22,23 In further regard to degradation pathways, we detected diminished levels of Cdm6pr in HFD fed mice, which suggests an impairment of lysosomal degrading pathway.24 These findings suggest a disruption of the lysosomal degradation pathway of proteins in HFD obese mice. Moreover, it has been reported that DIO mice present defects in the ubiquitin-proteasome system,20 as HFD obese mice presenting inflammation-induced protein aggregates in the hypothalamus were also unable to regulate body energy homeostasis.
Hierarchical-cluster analyses have been used as a multivariate approach for behavior and phenotype group recognition. It is important to highlight that in this study, different groups were assumed after D index analysis, thus being recognized as non-arbitrarily clustered groups. Therefore, we found three heterogeneous phenotypes of transcript expression in the hypothalamus for all mice analyzed. Regarding animal distribution among the three groups, the StD fed mice group was represented by at least one individual in the three distinct phenotype clusters, while all HFD fed mice were clustered into just one phenotype cluster. This finding may suggest a dynamic variation of transcript levels in a normal physiological process, as observed for the StD fed group, which could be ablated during the weight gain process in HFD fed mice.
In conclusion, our findings suggest that protein-sorting machinery may be altered in HFD fed C57BL/6j mice and may be associated with weight gain and hyperglycemia.
The study was supported by FAPEMIG grant number APQ-01996-14.
The authors declare that they have no conflict of interest.
All authors contributed to the preparation of the manuscript. FVMR, KSF, FSJ and AAB accomplished experimental design and literature analysis. FVMR, LAF, MLF, CMJ, CJCJ and KSF performed experimental assays and data analysis. FVMR, KSF, CJCJ and AAB wrote the manuscript.

©2018 Rubatino, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.