eISSN: 2575-906X


Research Article Volume 2 Issue 6
University of Antananarivo, Madagascar
Correspondence: Oliarinony Ranalison, University of Antananarivo, PB 906 Antananarivo (101), Madagascar
Received: October 09, 2018 | Published: November 20, 2018
Citation: Rakotomamonjy L, Oliarinony R. Phylogenetic, morphological and systematic studies of three genera of Cichlidae (Teleosts fish, Perciformes) endemic of Madagascar. Biodiversity Int J. 2018;2(6):538-543. DOI: 10.15406/bij.2018.02.00110
The aim of this research is to know the degree of reliability of the classification method based on molecular studies versus those based on the morphological characters. The hypothesis is as follows: the results from these two methods would be different. Eleven species belonging to the three genera of endemic Cichlids emerge from the genetic study using a mitochondrial DNA fragment COI, and four of which are new species for Science: Paratilapia sp. Nosy Be, Paratilapia sp. smallspots Est, Ptychochromis sp. Nosy be and Paretroplus sp. Lokoho. Morphological studies of 45 characters from 42 specimens did not correctly classify the three genera. Molecular systematics is thus a more reliable and informative method for studying the evolution of a given taxon and the link between its members.
Keywords: IOC DNA, Cichlidae, Madagascar, morphometry, phylogeny, systematic
Malagasy continental waters are recognized for their richness in biodiversity.1 Up to 41% of Madagascar's freshwater fish species (71 out of 172) are endemic, with two endemic families.2 Despite the fact that Madagascar is one of the conservation priority countries,3 its freshwater fish are still subject to several pressures, such as the introduction of carnivorous fish species, the destruction of forests , overfishing, habitat transformation for other human uses, and lack of knowledge about fish.4 Available information on the island's native freshwater fish is very low, although it is extremely threatened.5 Most research on these Malagasy freshwater fish is now outdated.5,6 Malagasy fish have been poorly studied because of the difficulty of conducting sampling.7 In short, minimal information and current knowledge on the systematics and evolution of freshwater fish from Madagascar exists, including some genera of Cichlids. The family Cichlids is one of the richest species among all vertebrates.8 This family is mainly found in Central Africa, Latin America, Madagascar and some native species in India and the Middle East.9 The Cichlidae family forms a monophyletic group and is subdivided into four subfamilies: Etroplinae (Indian and Madagascar), Ptychochrominae (Madagascar), Cichlinae (Neotropical Region) and Pseudocrenilabrinae (African), according to the classification derived from molecular and morphological data.10
However, the pressures on these genres continue to grow. Knowledge of diversity, in particular species richness, is a basic element to better understand biodiversity and its conservation. The present work, which focuses on morphological and genetic studies of some Cichlid species endemic to Madagascar, intends to enrich the knowledge of Malagasy ichthyological fauna.
The hypothesis of this research is as follows: "The classification resulting from the genetic study would be different from that resulting from the morphological study". Many authors such as Dayrat11 have proposed an integrative approach to taxonomic study. It is generally used to distinguish between cryptic species or to interpret an intraspecific variation. The main objective is to know the degree of reliability of the method of classification of the species operating the external characters (morphological and phenotypic) compared to the method of genetic classification; or (1) to know the phylogenetic relationships of the species belonging to the three endemic genera of Cichlidae of Madagascar: Paratilapia, Ptychochromis and Paretroplus, (2) to draw up the classification based on the external characters, (3) to identify the similarity and/or the difference results from the two classification processes; and (4) determine the main morphological characters that distinguish the groups studied.
Construction of the genetic tree
A small portion (corresponding to 1g) of the pectoral fin of each individual of the studied species each kept in a tube containing 90º alcohol serves as a genetic sample for the molecular study.
Total genomic DNA was extracted from tissue samples using proteinase K digestion at a concentration of 10mg/ml, followed by a standard salt extraction protocol according to Bruford et al.12 A mitochondrial DNA fragment of approximately 700 base pairs of Cytochrome Oxidase I (COI) was amplified using the primers developed by Hebert et al.,13 Ward et al.14 and Ivanova et al.15,16 PCR (Polymerase Chain Reaction) products were separated on an automated ABI 3130XL DNA sequencer at the Technical University of Braunschweig in 2012 and ABI 3730 at the Ludwig-Maximilians University of Munich in 2013.
The CLC Main Workbench 6.8.2 software was used for quality control of the DNA sequences and the sequences were aligned using the MEGA or Molecular Evolutionary Genetics Analysis software version 6.0.17
A search for the identity of all sequences studied and aligned was carried out in "GenBank". An NCBI nucleotide blast online search (http://www.ncbi.nlm.nih.gov/.) followed by an insertion of the sequence to be analyzed was then performed. The result of this search gives the species closest to that analyzed with a precision of the similarity rate between them; the sequence of this one was used for the construction of the phylogenetic tree. A total of 9 GenBank sequences were added to the 42 identified DNA sequences. The fasta file of the result of the MEGA alignment is opened with the software j-Model test 2.1.7. Its principle is to have a ΔAIC equal to 0.18 The Mr Bayes 3.2 software is then used to obtain a consensus tree, with a parameterization of 2 million generations (bootstrap method), a sampling frequency equal to 100 and an ignorance rate of 0, 25. The final result obtained from Mr Bayes, in the form of a nexus file, was opened with the Figtree software version 1.4.2., in order to know the degree of link existing between the species studied is known.
Classification based on external characters
The Dyer & Chernoff19 method was applied to the entire body of captured fish specimens. The choice of characters to be observed was based on the characteristics of the family of Cichlids proposed by Paugy et al.20 In total, 45 characters were studied. An encoding of each character state has been done, taking into account the character state that is primitive. The latter has been coded with 0, and the coding evolves according to the derivation of the state of the characters. The matrix thus obtained has been processed in a specific online software (http://www.genomes.urv.cat/UPGMA/index.php.), the DendroUPGMA (Vallve et al., 1999). For this study, the coefficient used is the RMSD or Root-Mean-Square-Distance to evaluate the statistical significance of the difference between two values. The resulting bit was inserted later in FigTree version 1.4.2.
Identification of morphometric and meristic differences
Measurement and counting: the 27 parameters to be measured refer to those proposed by Paugy et al.20 for studies of Cichlids, then adapted according to the needs of the author: 1-diameter of the eye (dO), 2-height of the anal fin (HA), 3-height of the body (HC), 4-dorsal fin height (HD), 5-pectoral fin height (HPc), 6-height caudal peduncle (HPC), 7-pelvic fin height (HPi), 8-head height ( Ht), 9-length of the anal fin base (LA), 10-mouth length (LB), 11-length of the dorsal fin base (LD), 12-length of the fin base pectoral (LPc), 13-length caudal peduncle (LPC), 14-pelvic fin base (LPi) length, 15-postorbital length (LposO), 16-length preanal (LPreA), 17-length predorsal (LPreD), 18-length pre-orbital(LPreO), 19-length prepectoral(LPrePc), 20-lengthprepelvic (LPrePi), 21-standardlength (LS), 22-totallength (LT), 23-length of the head (Lt), 24-mouth width (lB), 25-body width (lC), 26-lar interorbital gland (IO), 27-width of the head (lt ).
Measurements on the lateral face and in front view were made using a calipers D20TN Mitutoyo 200mm in length and the accuracy is 0.02mm.
Four distances between the insertions of the different fins were calculated after obtaining the values during the measurements: (a) the distance between the pelvic fin and the anal fin (dPiA); (b) the distance between the pectoral fin and the anal fin (dPcA); (c) the distance between the pectoral fin and the pelvic fin (dPcPi) and (d) the distance between the dorsal fin and the anal fin (dDA).
For this study, counting with the naked eye or a binocular loupe, fin (dorsal and anal) and scales along the longitudinal, lateral and operculum lines was performed.
Statistical analysis: statistical analyzes were performed on SPSS or Statistical Package for the Social Sciences version 17.00 to analyze the data from the study of morphometric and meristic parameters.
For this study the Mann-Whitney test21 was performed using relative values for morphometric parameters (variable parameters as a function of age) and exact values for meristic characters. PCA22 was used to describe the variables characterizing each individual. It makes possible to combine the variables studied in the form of factors (reduction in the number of variables) and to extract all the factors by keeping only those (containing the principal components) which are the most influential. Twenty localities have been collecting endemic Cichlid specimens from 2011 to 2014.
Molecular systematics
The result of the genetic analysis of the 51 COI mitochondrial DNA sequences of which 42 obtained in this study and nine, downloaded from GenBank, each containing 572 bases (sites), can be summarized by the phylogenetic tree with a standard average value of the frequency equal to 0.004496 (Figure 1). Pachypanchax omanolotus (Aplocheilidae) collected in Lake Amparihibe served as an external species for the study.
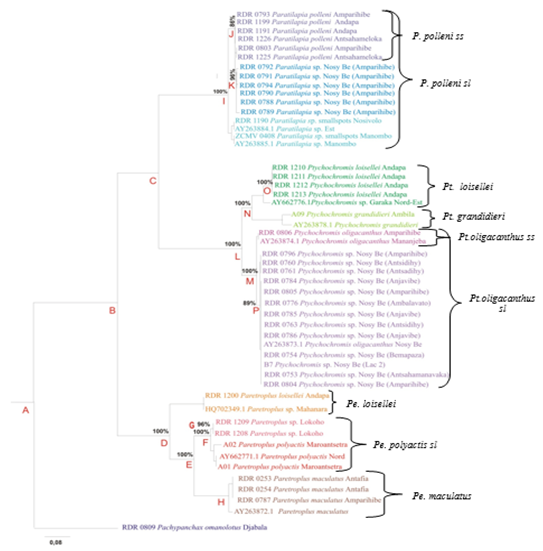
Figure 1 Phylogenetic tree of 11 endemic species of Malagasy Cichlids. RDM Sequences of Dr. Roger Daniel Randrianiaina; sequences A01, A02, A09, B7 and ZCMV of Prof Dr Miguel Vences; AY and HQ sequences from GenBank.
Paratilapia: a monophyletic genus, is thus a sister genus of Ptychochromis. Paratilapia sp. smallspots paired with Paratilapia sp. East and Paratilapia sp. Manombo is a sister species to the other two species of Paratilapia. Thus the smallspots species of the East is different from the largepots species of the North. For northern species, individuals Paratilapia sp. Nosy be and those of Paratilapia polleniss form two populations of different species, a difference maintained at 86%.
Ptychochromis: is a monophyletic genus. Ptychochromis oligacanthus sl is a sister species of the other two Ptychochromis loisellei and Ptychochromis grandidieri combined. Ptychochromis oligacanthus sl is composed of populations of two different species: Ptychochromis oligacanthus ss and Ptychochromis sp. Nosy Be with a sustained difference of 89% in P.
Paretroplus: is also a monophyletic genus. Paretroplus loisellei is a sister species of the other three Paretroplus species (Paretroplus polyactis, Paretroplus sp, Lokoho and Paretroplus maculatus). Two species of Paretroplus polyactis sl, different to 100% (in F), result from this study: Paretroplus sp. Lokoho and Paretroplus polyactis Maroantsetra. The latter is paired with Paretroplus polyactis North of the "GenBank".
Morphological systematics
A total of 45 external characters were studied on each of the 42 fish specimens and obtained the dendrogram of Figure 2. The genera Ptychochromis and Paretropluss are sister groups and these two groups combined form a sister group of the genus Paratilapia.
Paratilapia: is a monophyletic group and Paratilapia polleni sl is a cryptic species. Antsahameloka harbor two types of populations of Paratilapia polleniss with different characters (polymorphic); and the population of Paratilapia sp. Nosy Be (Amparihibe), in spite of their genetic difference with that of Paratilapia polleniss Amparihibe, presents a great similarity of the characters studied.
Ptychochromis: belongs to a monophyletic group and is a cryptic genus. The four species are not monophyletic groups. Individuals of Ptychochromis grandidieri Amboditandroho form a sister group of all other individuals of the genus Ptychochromis (in E). Thus, the individuals of Ptychochromis grandidieri differ in their characters. Also, referring to node N, Ptychochromis sp. Nosy Be and Ptychochromis oligacanthus ss Amparihibe (Nosy Be) show morphological differences. The individual of Pt. Loisellei RDR 1213 has characteristics that differentiate him from all other individuals belonging to this species.
Paretroplus: is a monophyletic group. It is the same for the species Paretroplus loisellei which is a sister species of the three others combined. Individuals of Paretroplus polyactis Pangalanes can be divided into several batches of individuals with different characters; it is the same for Paretroplus maculatus Antafia. Thus, Paretroplus polyactis and Paretroplus maculatus are both paraphyletic groups (in J).
Inter and intra group differences
According to the results of the U test carried out for the genera Ptychochromis and Paratilapia with a number of individuals of Ptychochromis n1=15, and that of Paratilapia n2=11 and for α=0.05, the value of Utable=44. In total, 23 parameters out of the 37 studied make it possible to distinguish the two kinds, a 65% difference. Thus, Paratilapia is distinguished from Ptychochromis by these main characters: a larger body, a larger head, a longer and wider anal fin, a longer caudal peduncle and fewer rays of the dorsal fin.
For the second group compared, the genera Ptychochromis and Paretroplus, the numbers of the individuals are respectively n1=15 and n2=16 and for α=0.05, Utable=70. According to the statistical results, 15 characters out of the 37 studied are different between them. The body, the fins and the meristic characters are the main differentiations of these genera.
The result from the U test shows that for Paratilapia (at 11 individuals) and Paretroplus (at 16 individuals) and for α=0.05, Utable=47. Paratilapia differ of Paretroplus of 14 morphological characters out of the 37 studied. As for Paretroplus, it is distinguished from Paratilapia partly by: a higher body height, the rays of the dorsal fin and the anal fin more numerous and more scales on the lid and along the longitudinal and lateral lines higher.
The main morphometric and meristic differences of Paratilapia polleni ss and Paratilapia sp. Nosy Be remains especially in the insertion of the pelvic fin and the height of the dorsal dorsal.
Species of the genus Ptychochromis differ mainly on the characters of the head. Ptychochromis loisellei and Ptychochromis grandidieri show no significant difference.
For the genus Paretroplus, the difference varies according to the species compared: Paretroplus loisellei and Paretroplus polyactis are 84% different for the 37 parameters, whereas Paretroplus loisellei and Paretroplus maculatus show no significant difference (Figure 3).
The result of the PCA shows that three factors account for 89.546% of the 39 variables. But only two regression factors were selected according to their importance: factor 1: body shape, and factor 3: number of hard rays of the dorsal fin and number of scales along the longitudinal line and the upper lateral line.
Out of twenty localities collecting endemic Cichlid specimens for three years from 2011 to 2014. The following points are worthy of mention.
Identities of the studied species
Several studies have shown that the subfamily Etroplinae (including the genus Paretroplus) is sister to Ptychochrominae (composed in part by Paratilapia and Ptychochromis) plus African Cichlids and Neotropical.10,23‒25
Concerning the genus Paratilapia, this study made it possible to distinguish three species: P. polleni ss the typical form and two other new species: P. sp. Nosy Be (Amparihibe) and P. sp. smallspots Est. The first species, P. polleni ss or P. polleni "largepots" corresponds to that described by de Rham & Nourissat26 to name individuals with large points and having a distribution in the northern part of Madagascar. The characterization of this species as "sensu stricto" or typical is first justified by the Andapa specimens with large dots and which are of the same type as those of Sambirano;26 a locality where the holotype described by Bleeker in 1868 was captured. The second species, P. sp. Nosy Be having big points is according to the bibliography and the results of "GenBank", a species of P. polleni "largepots". However, the genetic tree from this study to see the links between species shows that this species differs from P. polleni ss to 86%. It should be noted that all these individuals were collected in Lake Amparihibe Nosy Be. In 1882, Sauvage27 described the species P. bleekeri, a species also having large dots but collected in Antananarivo. This description is described as "hasty" by many authors26,28 and is considered invalid even though Loiselle & Stiassny29 adopted this nomenclature. As a result, no other description after that of Bleeker has been made, thus the second species resulting from this study P. sp. Nosy Be is a still undescribed species and is a "new species". Lake Amparihibe is home to two different populations of Paratilapia "largepots" including the species P. sp. largepots Nosy Be who is absent in Andapa. The third species, P. spsmallspots East is considered another species after Rham & Nourissat,26 but as P. polleni East by "GenBank". The accuracy of the origin of this species is accurate considering that the places of collection of our specimens were the Eastern part of the Island (case of Nosivolo) and the South-East part (case of Manombo). Other authors30 confirm that it really occupies these localities. In 1878, Bleeker described the species Paracara typus East, which was later modified into Paratilapia typus31 and has a strong resemblance to P. sp. smallspots East but whose origin of the only exemplary type is unknown. Indeed, P. sp. smallspots East could be synonymous with that described by Bleeker or a new species that is not yet described.
For Ptychochromis, it is composed of four species: Pt. loisellei, Pt. grandidieri, Pt. oligacanthus and Pt. Sp. Nosy Be. Stiassny& Sparks32 described the species Pt. loisellei, formerly named Pt. Sp. Northeast Garaka. Pt. loisellei being genetically closer to Pt. grandidieri spreading also to the east, and further north. Pt. oligacanthus is so named because: the holotype that Bleeker described in 1868 came from Sambirano and even though our specimen was collected in Nosy Be, it is 100% identical to that of GenBank from Mananjeba, a very locality close to the typical locality, and whose two basins touch each other; furthermore, Sparks33 states that populations in the north possess the characters described by Bleeker.31 Apart from this first collection of the holotype, another from Ampobilava Lake (Nosy Be) was, at the same time, described by Bleeker.31 Sparks33 assumes that Pt. sp. Mananjeba and Pt. Sp. Nosy Be are of the same species but Loiselle34 finds a difference between the population of Nosy Be and that found on Grande Terre. He also adds that each lake in Nosy Be is populated by populations of different species that have diverged from a common species. As the molecular analysis shows a 100% difference between the Nosy Be and Mananjeba species and a 100% resemblance of the species Pt. sp. Nosy Be with that of Nosy Be. In addition, Loiselle34 discovered formally the non-validity of the specific name Pt. nossibeensis proposed by Guinane,35 this species Pt. sp. Nosy Be is then undescribed and is a "new species". Thus, this study made it possible to elucidate the doubts about the belonging of Ptychochromis Mananjeba and that of Nosy Be to the same species.26
Martinez & Sparks36 confirms the results on the links between species of the genus Paretroplus. Paretroplus polyactis sl and Paretroplus loisellei are the two species belonging to this genus, found in the eastern part of Madagascar. However, according to the phylogenetic tree obtained, Paretroplus maculatus is closer to Pe. polyactis. Thus, the statement that Pe. polyactis is the only genus occupying exclusively the eastern part and is in a clade apart from all other Paretroplus,7 has not been confirmed. Polyactis Maroantsetra and Pe. sp. Lokoho are, according to genetic studies, 100% different species despite their same distribution in the eastern part of Madagascar. According to the result of GenBank, Pe. Polyactis Maroantsetra is 100% identical to Pe. polyactis in the North. The explanation of the classification choice is that the holotype of Pe. polyactis described by Bleeker in 1879 comes from the Canal des Pangalanes. Loiselle,37 due to the difference in color, suspects the presence of 2 or more species Pe. polyactis. Thus, a probable speciation within this species would have occurred in this northeastern part of the island. As a result, the species Pe. sp. Lokoho is also a "new species" (Figure 4).

Figure 4 Species studied (Photos Randrianiaina, 2013 and Nourissat, 2004) (a) Paratilapia polleniss (b) Paratilapia sp. smallspots East. (c) Ptychochromis grandidieri. (d) Ptychochromis loisellei. (e) Ptychochromis oligacanthus ss -E: Scale. (f) Ptychochromis sp. Nosy Be. (g) Paretroplus sp. Lokoho. (h) Paretroplus maculatus. (i) Paretroplus loisellei. (j) Pachypanchax omanolotus.
Similarities and differences between molecular systematics and morphological classification
The three genera have been well separated on the dendrogram of characters and each possess their respective clade. Although the three genera are monophyletic groups, on the phylogram and statistically, the genera Ptychochromis and Paratilapia are more distant and have the most difference. This is opposed to molecular results. With this difference of results, the following hypothesis could be considered "the phenotypically similar characters can be genetically different" knowing that different codons can code for the same protein and all of these proteins are the phenotypic image reflecting the genotype of an individual. The fact that Ptychochromis and Paretroplus, likewise Paratilapia and Paretroplus, show a difference over almost all the meristic characters, would be a justification for the belonging of the two genera (Ptychochromis and Paratilapia) to the subfamily Ptychochrominae. In addition, Paretroplus found in the subfamily Etroplidae is distinguished from the other two genera by the height of its body and belongs to the clade of high bodies,38 by the possession of a number of hard rays the anal fin more than double that of the Ptychochrominae (which is still 3 according to Sparks,7) and also the dermal cover at the base of the scales of the dorsal and anal fins.26 Thus, compared to individuals of Paratilapia sp. Nosy Be, those P. polleni ss are more varied. This is justified by the result of the PCR which demonstrates that for the body form factor, the Andapa individuals are widely dispersed. Genetically, these varied individuals belong to two different species.
With regard to the genus Ptychochromis, Pt. lawellei and Pt. grandidieri being more genetically similar and morphologically identical (no significant difference in traits between these two species was observed in the Mann-Whitney test result). Stiassny & Sparks32 confirm that the similarity of these two species is very high. Finally, the two species Paretroplus maculatus and Pe. polyactis are more closely related genetically and on the phylogram. According to the ACP, the individuals of Pe. sp. Lokoho are isolated from those of Pe. polyactis, which confirms the result of the molecular study showing the belonging of these individuals to 2 different species.
Thus, if it is necessary to have an idea about the distributions at the genus level, the morphological classification is still instructive. But in the case where the details of the systematics, ie the degree of relations between taxa (at the genus and especially inter and intraspecific level) and the direction of evolution, the genetic method is reliable and more informative. Ambiguities or differences in morphological classification, especially for cryptic species, are solved by the genetic method with a precision of the relationship. This finding has also been observed for other classes of vertebrates.39
Also, the morphological differentiations do not necessarily translate into molecular speciations. In addition, although many informative biogeographic studies have been conducted many times, ambiguities about classification are not always resolved. Like the case of Paratilapia polleni, several more names are attributed to it and it is a complex species that needs a systematic revision.40 However, during this study, molecular genetics allowed to discover new species, a great asset that would not emerge from the morphological study. In sum, four (04) new species still undescribed spring from this phylogenetic study: Paratilapia sp. smallspots East, Paratilapia sp. largespots Nosy Be, Ptychochromis sp. Nosy Be and Paretoplus sp. Lokoho.
In short, the hypothesis of this study is accepted; a significant difference exists between molecular systematic and morphological classification. This difference has appeared from the genus level and is more accentuated at the species level with a confirmed reliability of the genetic method. The result demonstrates that the combination of genetic studies and phenotypic diversity provides robust information on intraspecific distribution that will subsequently be a fundamental element for future conservation projects.
For the Nosy Be Lakes, researchers believe that the large satellite island of Nosy Be being virgin of non-native predators, would be one of the highest conservation priorities.26,34 Conservation actions include projects for the rational management of lake waters, especially for Lake Amparihibe, many conspecific species are observed in this study. After this study, the observation of several endemic and new species occupying these lakes and subsequent research that deserve to be carried out are additional arguments for conserving these lacustrine ecosystems of Nosy Be.41,42
We extend our gratitude to the Volkswagen Foundation for funding this study. We thank Professor Noromalala RAMINOSOA, Dr. Roger Daniel RANDRIANIANA and Dr. Fanomezana RATSOAVINA for their invaluable help. We also thank Professor Dr. Miguel VENCES for giving us the sequences of the fish he has collected to complete this study and Mr. Guy TAM HYOK for having agreed to collaborate during the collection of samples in the APPA center in Andapa, Madagascar.
None.
The authors declare that there is no conflict of interest regarding the publication of this article.

©2018 Rakotomamonjy, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.