Advances in
eISSN: 2377-4290


Image Article Volume 9 Issue 4
1Department of Pathology, Maimonides University, Argentina
2Department of Pathology, University of Buenos Aires, Argentina
Correspondence: Jorge Oscar Zarate, Department of Pathology, Digital Pathology Laboratory, Faculty of Medicine, Maimonides University, Argentina
Received: July 19, 2019 | Published: July 22, 2019
Citation: Zarate JO, Zárate Vidal JM. Optical digital biopsy; the phisical photo of pigmentary epithelium. Adv Ophthalmol Vis Sys. 2019;9(4):97-98. DOI: 10.15406/aovs.2019.09.00354
In the secuentiation of image by pixelometry of isolated cells, studing the pigmentary epithelium of retina we can appreciate three areas: the nucleus and the cytoplasm corresponding to cells of the pigmentary epithelium, physical, alive, in situ, achieved through the sequencing of optical coherence tomography (OCT) images (Figure 1).1–16 The steps for this determination can be summarized in:
This procedure aims to adapt the image for later analysis. Therefore, the following operations have been carried out:
This procedure has allowed us to: first extract a band to be able to then operate on it, this is convenient as it is necessary within the process convert the image to binary image. Then, soften the image using a median filter; this because the totality of the detail that contains the image, since they behave as noise for the end pursued. And finally the image has been rotated in order to place it in its better angle, in such a way, that the extraction of the region of interest contains pixels that represent it optimally. The adequate selection of the region of interest allows operating only on the pixels that contain relevant information.
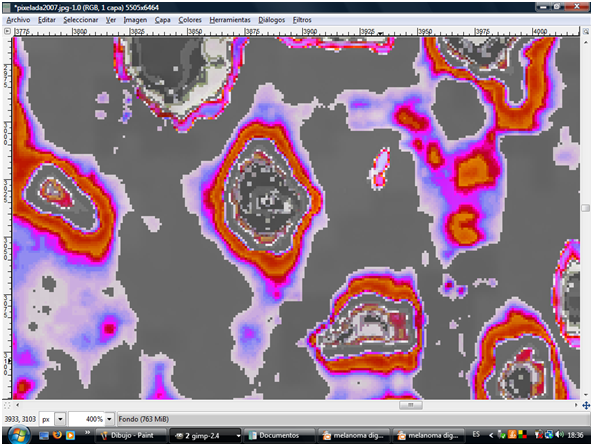
Figure 1 Physical cells of the pigment epithelium of the retina. Photo obtained from an OCT after the sequencing of images, that is, without an invasive component.
Obtaining the required parameters for work on the image has been made by segmenting the image. This procedure has allowed us to subdivide the images into their constituent objects: nucleus and cytoplasm, taking advantage of the fact that the gray levels of the nucleus and the cytoplasm are differentiable. The segmentation of the images was done by the thresholding method, using information provided by the histograms of the levels of gray of the processed images and the corresponding averages, as
it shows in the following diagram:
The determination of the number of pixels represented by the cell as a whole, It was done using as a threshold the average of the total pixels of the image. This it has allowed us to label each pixel as belonging to the background or the cell. To obtain the pixels corresponding to the nucleus, the threshold was used gray level values corresponding to the left inflection points and right of the curve to the left of the histogram; and, for the pixels corresponding to the cytoplasm, the left and right inflexion points of the curve to the right of the histogram. The methodology is repeatedly pixelometric, pixelográfica and could be added pixeloarquitectural. Each binary image, which taken to the subcellular level can be a protein has a code based on pixels, with so many probabilities that it exceeds the genome.
None.
Author declares that there are no conflicts of interest.

©2019 Zarate, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.