MOJ
eISSN: 2379-6294


Research Article Volume 4 Issue 4
1Hachinohe National College of Technology, Japan
2Himeji Dokkyo University, Japan
Correspondence: Yu F Sasaki, Toxicology Laboratory, Taisho Pharmaceutical Co., Ltd., Yoshino-cho 1-403, Saitama-shi, Saitama 330-8530, Japan, Tel 8117 8277 296
Received: July 04, 2018 | Published: July 12, 2018
Citation: Kawaguchi S, Nakamura T, Tsuda S, et al. Detection of in vitro genotoxicity of pro-mutagens using the comet assay under human and rat liver S9 fractions. MOJ Toxicol. 2018;4(4):255–261. DOI: 10.15406/mojt.2018.04.00109
The metabolic activating ability of human liver S9, human lung S9 (HLuSP-E), and rat liver S9 were compared. One of the human liver S9 (HLSP-E) was prepared from a pool from 15 donors and the other (HLS-014E) from a liver sample that had high levels of P450 activities. Rat liver S9 was prepared from male Sprague–Dawley rats either untreated (RLSP-E) or pretreated with phenobarbital/5,6-benzoflavone (RL-PB/BF). Human lymphoma WTK1 cells were treated with 37 pro-mutagens that required cytochrome P450 or NADPH-P450 reductase for metabolic activation and their genotoxicity was determined by the comet assay. HLS-014E and RL-PB/BF had a higher P450 enzyme activity and activated a greater variety of pro-mutagens than others. The results of the human liver S9 and RL-PB/BF were discrepant for only 2 out of 37 pro-mutagens; phenacetin was positive with human liver S9 but not with RL-PB/BF, and benz[a]anthrathene was positive with RL-PB/BF but not with human liver S9. Both RL-PB/BF and human liver S9 activated pyrene, IQ, MeIQ, 1-nitropyrene, and N-nitrosopiperidine while RLSP-E did not. In spite of the existence of species differences in CYP isoforms and differences of CYP activity, therefore, there were no appreciable qualitative differences in pro-mutagen activating ability of the human liver S9 and RL-PB/BF, suggesting the possibility that the genotoxicity of some pro-mutagens that are genotoxic with human liver S9 are missed by the use of liver S9 from rats that were not pretreated to induce CYP. Therefore, conventionally used CYP-induced rat liver S9 fraction is useful for the evaluation of the genotoxic risk for humans in spite of the species difference in CYP isoforms.
Keywords: rat liver S9, human liver S9, metabolic activation, comet assay
Genotoxicity tests are used to detect mutagens and carcinogens, study the mechanisms of chemical mutagenesis and carcinogenesis, and assess the genotoxic and carcinogenic hazards of chemicals to humans. Genotoxicity tests also play a critical role in drug development. For all those uses, data is usually obtained from experimental organisms and extrapolated to humans. Pro-mutagens are inactive until enzymes transform them to electrophilic species capable of covalently binding to DNA, so metabolic activation is a critical step in mutagenesis.1 Metabolic activation systems are necessary for testing the potential genotoxicity of pro-mutagens. Cytochrome P450 (CYP) in liver S9 fractions plays an important role in activating pro-mutagens to proximate and/or ultimate mutagens.1,2 Liver S9 prepared from rats pretreated with phenobarbital/5,6-benzoflavone (PB/BF) or Aroclor 1254 is routinely used for that purpose in genotoxicity tests because they are simple, convenient and cost-effective.2,3 Human liver S9, however, are known to behave differently than rat liver S9 in the activation of four compounds and cigarette smoke condensate.4
The liver P450 involved in the activation of some chemical carcinogens has different isoforms in humans and rats.1 Although knowing the difference in their activity is important for an accurate extrapolation of genotoxicity test data to humans, the number of pro-mutagens tested with human liver S9 is too small for a definitive conclusion.5 Using the Ames test, Hakura et al.,5 compared the genotoxicity of 13 pro-mutagens activated with liver S9 from 4 humans (H3, H8, H12, and H14), from rats treated with PB/BF (RL- PB/BF), or from un-treated rats (RL). They found that (1) the S9 fractions differed in magnitude of pro-mutagen activation, (2) three of the human preparations showed equivalent activity while the fourth showed higher activity, (3) with 10 pro-mutagens, the order of magnitude was RL-PB/BF>H14>RL>H3, H8, and H12, while with 2-aminoanthracene (2-AA), N-nitrosodimethylamine (DMN), and 1-nitropyrene (1-NP), the order was H3, H8, H12, and H14 > RL >RL-PB/BF. Based on the data, the authors concluded that the human liver S9 fraction is much more useful than the rat’s in the Ames test S9 for evaluation of genotoxicity on humans.5 Because of species differences in CYP isoforms, we agree that the use of a human liver S9 fraction is important in genotoxicity tests that assess risks to humans. However, before concluding that human liver S9 is more useful than rat liver S9, many matters must be resolved: e.g., (1) whether we can obtain S9 human liver that is representative of the general population, which shows great individual differences in CYP activity based on genetic and life-style, and (2) whether many pro-mutagens can be activated by human but not rat S9. According to the experiment by Hakura et al.,5 activation of 13 pro-mutagens varied quantitatively depending on the S9 fraction and compound, but no pro-mutagens were activated only by human S9. Therefore, it is too early to conclude that there is an advantage of using human, rather than rat, S9 in genotoxicity tests.
We undertook the present study to ascertain whether there are appreciable differences between the pro-mutagen activating ability of human and rat liver S9. Although the Ames test is widely used for detecting the genotoxicity of chemicals,2 there are some discrepancies between genotoxicity in microorganisms and cultured mammalian cells. For example, IQ, MeIQ, and diMeIQx do not induce chromosome aberrations in,6,7,8 and PhIP shows only clastogenic activity in, excision repair-deficient Chinese hamster cells,7 but all are potent bacterial mutagens.9,10 In this study, we tested the genotoxicity on cultured human cells of 40 pro-mutagens that are preferentially activated by different P450 isozymes, and we used the comet assay to assess DNA damage. The pro-mutagens we used included polycyclic aromatic hydrocarbons, aromatic amines, heterocyclic aromatic amines, N-nitrosoamines, nitropyrene, and others.
Chemicals, cells and medium
Table 1 lists the pro-mutagens tested, their abbreviations, CAS numbers, while Table 2 lists the P450 isoforms preferentially involved in their activation. Table 3 lists the 5 kinds of S9 fractions we used, along with the CYP concentration and activity. Human S9 fractions were prepared from samples of non-transplantable livers and lungs of donors procured from the National Disease Research Interchange (NDRI) in Philadelphia, PA, with permission for research use, based on the international partnership between the NDRI and the Human and Animal Bridge Discussion Group (HAB) in Japan. Pooled human liver S9 fractions were prepared from tissue from 15 donors that is kept by the HAB. HLS-014E (which is the same S9 (H14) used by Hakura et al.,5 showed the highest P450 activity of the 18 human liver tissues stocked by the HAB, possibly due to induction by anti-asthma agents for 10 years.5 Pooled human lung S9 were prepared from tissue from donors that is kept by the HAB. Rat S9 fractions were prepared from livers of male SD rats that had been treated with phenobarbital and 5,6-benzoflavone (CYP-induced) or and untreated. Theformer were obtained from the Oriental Yeast Co. Ltd. and the latter from the HAB. The human lymphoma (WTK1) cells, which were obtained from Dr. Masamitsu Honma, National Institute of Hygienic Sciences, Tokyo, were grown in a RPMI1640 medium (Nissui Pharmaceutical Inc., Tokyo) supplemented with 10% fetal bovine serum (HyClone Laboratories, Inc., U.S.A.). The cells were grown at 37°C in a humidified atmosphere containing 5% CO2.
Treatment
We treated logarithmic growth phase cells (1x106cells/ml treatment medium) with each chemical and vehicle alone at 37°C for 4h in the treatment medium containing each S9 fraction. Cells were treated at the range at which each chemical was comet assay-negative in the absence of S9 mix and gross precipitation was not observed in the treatment medium. For pro-mutagens not showing any cytotoxicity to WTK1 cells, the highest treatment concentration was 5000µg/ml. For pro-mutagens showing cytotoxicity to WTK1 cells, the highest treatment concentration was the concentration at which cell viability (measured by the trypan blue exclusion test) immediately after treatment was about 70–80%. The treatment medium contained 4mM MgCl2 6H2O, 16.5mM KCl, 2.5mM glucose-6-phosphate, 2mM NADPH, 2mM NADH, 50mM Na2HPO4, 50mM NaH2PO4, and 5% S9 fraction. Cells were subjected to the comet assay sampled immediately after chemical treatment.
Comet assay
Seventy fiveµl of 1% agarose GP-42 (Nakalai Tesque, Inc., Kyoto, Japan) was quickly layered on a slide (Matsunami Glass Ind., Ltd., Osaka, Japan) and covered with another slide glass. The treated cells were suspended in 1% agarose-LGT (Nakalai Tesque, Inc.) and 75µl of cell suspension (5x105cells/75µl) was quickly layered at the same manner after removing the slide glass. Finally, 75µl of 1% agarose GP-42 was quickly laid on again. The slides were lysed immediately in a solution (pH 10) of 2.5MNaCl, 100mM Na2EDTA, 10mM Trizma, 1% sarkosyl, 10% DMSO, and 1% Triton X-100 at 4°C for 60min.11 The slides were placed on a horizontal gel electrophoresis platform, and covered with an alkaline solution made up of 300mM NaOH and 1mM Na2EDTA (pH>13). The slides were left in the solution for 20 min to allow the unwinding of the DNA and expression of alkali-labile sites. The power supply was set at 25V and 250mA. The DNA was electrophoresed for 20min and the slides were rinsed with 400mM Trizma (pH 7.5) to neutralize the excess alkali. Each slide was stained with 50µl of 20µg/ml ethidium bromide (Wako Pure Chemical Industries, Ltd.) and covered with a coverslip.
The cells on one slide were photographed (black and white 400 ASA Fuji film) as seen with the aid of a fluorescence microscope (Olympus at 200x magnification) equipped with an excitation filter of 515-560nm and a barrier filter of 590 nm. The length of the comet tail was measured for 50 cells per treatment group. The relationship between chemical treatment and migration was analyzed by ANOVA and the Dunnett test. Results
Figures 1-5 shows the length of the comet tail for each pro-mutagen in the presence of five kinds of S9, and without any metabolic system. Experimental data are summarized qualitatively in Figures 6-10. At the selected pro-mutagen concentrations, cell viability was never lower than 70% (data are not shown) and any pro-mutagens did not increase tail length without any metabolic system. With CYP-induced rat liver S9 (RL-PB/BF), all the studied pro-mutagens, except phenacetin, increased tail length significantly. Under RLSP-E, pyrene, IQ, MeIQ, and 1-NP, in addition to phenacetin, did not increase tail length significantly (Figure 6). Phenacetin, in contrast, yielded increased tail length in the presence of pooled human liver S9 HLSP-E (Figure 7). With HLSP-E, however, three pro-mutagens (B[a]A, 1-AP, and 1-NN) showing increased tail length with RL-PB/BF did not show (Figure 8). With HLSP-E or HLSP-014E, however, two pro-mutagens (1-AP, and 1-NN) out of those five showed increased tail length (Figure 7). Although CYP1A2, CYP2D6, and CYP3A4 were more than twice as active in HLS-014E than in HLSP-E (Table 3), five pro-mutagens (pyrene, B[a]P, 2-AA, IQ, and MeIQ) increased tail length with HLSP-E but not with HLS-014E (Figure 9). On the other hand, only two (1-AP and 1-NN) increased tail length with HLSP-014E but not with HLSP-E (Figure 9). The activity of most CYPs were far lower in human lung S9 (HLuSP-E) than human liver S9 (HLS-014E and HLSP-E) (Table 3). Although many pro-mutagens increased tail length with the human liver S9 but not the human lung S9, 1-AP and pyrene increased tail length with the human lung S9 but not with the human liver S9 (Figure 10).
Pro-mutagens |
Abbreviation |
CAS number |
Sourcea |
Vehicleb |
| Aromatic hydrocarbons |
|
|
|
|
| Benzene |
|
71-43-2 |
W |
D |
Benz[a]anthrathene |
B[a]A |
56-55-3 |
T |
D |
Benzo[a]pyrene |
B[a]P |
50-32-8 |
S |
D |
Phenanthrene |
|
85-01-8 |
T |
D |
Pyrene |
|
129-00-0 |
T |
D |
| Aromatic amines |
|
|
|
|
2-Aminoanthracene |
2-AA |
613-13-8 |
W |
D |
Aniline |
|
62-53-3 |
W |
D |
2,4-Diaminotoluene |
2,4-DAT |
95-80-7 |
W |
D |
1-Naphthylamine |
1-NA |
134-32-7 |
T |
D |
2-Naphthylamine |
2-NA |
91-59-8 |
* |
D |
Phenacetin |
|
62-44-2 |
W |
D |
1-Aminopyrene |
1-AP |
1605-67-3 |
W |
D |
Benzidine |
|
92-87-5 |
* |
D |
3,3’-Dichlorobenzidine |
DCB |
91-94-1 |
S |
D |
o-Tolidine |
|
119-93-7 |
T |
D |
Heterocyclic amincs |
|
|
|
|
Trp-P-1 acetate |
|
68808-54-8 |
W |
S |
Trp-P-2 acetate |
|
62450-10-3 |
W |
S |
AC |
|
26148-68-5 |
W |
D |
IQ |
|
76180-96-6 |
W |
D |
MeIQ |
|
77094-11-2 |
W |
D |
PhIP hydrochloride |
|
105650-23-5 |
W |
S |
Dialkyl N-nitrosoamines |
|
|
|
|
N-Nitrosodimethylamine |
DMN |
62-75-9 |
W |
S |
N-Nitrosoethylmethylamine |
EMN |
10595-95-6 |
W |
S |
N-Nitrosodiethyamine |
DEN |
55-18-5 |
W |
S |
N-Nitrosodiethanolamine |
DEolN |
1116-54-7 |
S |
S |
N-Nitrosodipropylamine |
DPN |
621-64-7 |
N |
D |
N-Nitrosodibutylamine |
DBN |
924-16-3 |
S |
D |
N-Nitrosomorpholine |
NMOR |
59-89-2 |
S |
S |
N-Nitrosopiperidine |
NPIP |
100-75-4 |
Ka |
S |
N-Nitrosopyrrolidine |
NPYR |
930-55-2 |
Ka |
D |
Azo dyes |
|
|
|
|
Azobenzene |
|
103-33-3 |
T |
D |
p-Aminoazobenzene |
pAAB |
60-09-3 |
W |
D |
p-Dimethylaminoazobenzene |
DAB |
60-11-7 |
W |
D |
|
|
|
|
|
Nitro compounds |
|
|
|
|
1-Nitronaphtherene |
1-NN |
86-57-7 |
T |
D |
1-Nitropyrene |
1-NP |
5522-43-0 |
T |
D |
|
|
|
|
|
Others |
|
|
|
|
Aflatoxin B1 |
AFB |
1162-65-8 |
W |
S |
Cyclophosphamide |
CYP |
50-18-0 |
W |
S |
Table 1 Pro-mutagens tested in this study
aK: Kanto Chemical Co., Inc., Tokyo (Japan); Ka: Katayama Chemical; S: Sigma Chemical Co., St. Louis, MO (U.S.A.). *2-Naphthylamine was synthethized from 2-naphththol and ammonium sulfite in Laboratory of Genotoxicity, Hachinohe National College of Technology, Aomori, Japan. Benzidine was synthethized from hydrazobenzene in Laboratory of Genotoxicity, Hachinohe National College of Technology.
bD, DMSO; S, Saline
Enzymes |
Pro-mutagens |
CYP1A1 |
B[a]P and other polycyclic hydrocarbons, DBN |
CYP1A2 |
Aromatic amines, Aromatic amides |
CYP2A6 |
Phenacetin, DEN, DPNa, NMORa, NPIPa, NPYRa |
CYP2C9 |
Polycyclic hydrocarbons |
CYP2E1 |
Benzene, Aniline, DMN, EMNa, DEolNb, Styrene, Acrylonitrile |
CYP3A4 |
Polycyclic hydrocarbon dihydrodiols, 6-Aminochrysene, 1-NP, AFB1 |
NADPH-P450 reductase |
Azo compounds, Nitro compounds |
Table 2 Enzymes primarily involved in metabolic activation of specific pro-mutagens
References for enzymes primarily involved in metabolic activation:13-15
Source of |
Protein1 |
CYP concentration (pmol/mg protein)2 |
|||||
S9 fraction |
(mg/ml) |
CYP1A2 |
CYP2C9 |
CYP2C19 |
CYP2D6 |
CYP2E1 |
CYP3A4 |
HLSP-E |
21 |
11.6 |
NT |
NT |
7.01 |
56.1 |
46.0 |
HLS-014E |
20 |
33.2 |
NT |
NT |
15.0 |
35.3 |
101 |
HLuSP-E |
20 |
2.76 |
NT |
NT |
ND |
ND |
ND |
RLSP-E |
20 |
18.6 |
NT |
NT |
49.4 |
65.8 |
19.0 |
RL-PB/BF |
20 |
95.2 |
NT |
NT |
1.20 |
54.6 |
11695 |
Source of |
|
Enzyme activity (pmol/min/mg protein) |
|||||
S9 fraction |
|
CYP1A23 |
CYP2C94 |
CYP2C195 |
CYP2D6 |
CYP2E17 |
CYP3A48 |
HLSP-E |
|
32.89 |
13.34 |
1.17 |
7.46 |
1041.2 |
995.1 |
HLS-014E |
|
77.84 |
38 |
1.18 |
18.43 |
973.3 |
4383.3 |
HLuSP-E |
|
2.695 |
0.0077 |
1.644 |
ND |
ND |
ND |
RLSP-E |
|
90.5 |
0.319 |
7.96 |
529.6 |
952.9 |
1214.8 |
RL-PB/BF |
|
1655 |
127.2 |
ND |
81.3 |
2521.6 |
3422.2 |
Table 3 CYP concentration and activity in human and rat organ S9 fractions
NT: not tested; ND: not detected
1Protein concentration of liver microsome measured by Bradford pigment-binding method. 2Measured by the carbon monooxide-differential spectrum method developed by Omura and Sato. 3O-deethylation activity of ethoxyresorufin. 4Methylhydroxylation activity of tolbutamide. 54’ -Hydroxylation activity of S-mephenytoin. 64-Hydroxylation activity of debrisoquine. 76-Hydroxylation activity of chlorzoxazone. 83-Hydroxylation activity of diazepam.
Data were obtained from HAB.

Figure 1 Comet assay with with aromatic hydrocarbons in the presence of human and rat tissue S9s and a control. Comet slides were prepared immediately after the exposure to each aromatic hydrocarbons for 4h. The error bars indicate standard deviation of the mean of three independent trials. *Significant difference from untreated control: p<0.05.

Figure 2 Comet assay with aromatic amines in the presence of human and rat tissue S9s and a control. Comet slides were prepared immediately after the exposure to each aromatic amines for 4h. The error bars indicate standard deviation of the mean of three independent trials. *Significant difference from untreated control: p<0.05.

Figure 3 Comet assay with heterocyclic amincs in the presence of human and rat tissue S9s and a control. Comet slides were prepared immediately after the exposure to each heterocyclic amincs for 4 h. The error bars indicate standard deviation of the mean of three independent trials. *Significant difference from untreated control: p<0.05.

Figure 4 Comet assay with dialkyl N-nitrosoamines and azo dyes in the presence of human and rat tissue S9s and a control. Comet slides were prepared immediately after the exposure to each dialkyl N-nitrosoamines and azo dyes for 4h. The error bars indicate standard deviation of the mean of three independent trials. *Significant difference from untreated control: p<0.05.
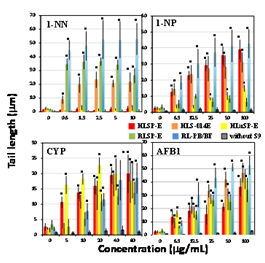
Figure 5 Comet assay with nitro compounds and other pro-mutagens in the presence of human and rat tissue S9s and a control. Comet slides were prepared immediately after the exposure to each nitro compounds and other pro-mutagens for 4h. The error bars indicate standard deviation of the mean of three independent trials. *Significant difference from untreated control: p<0.05.
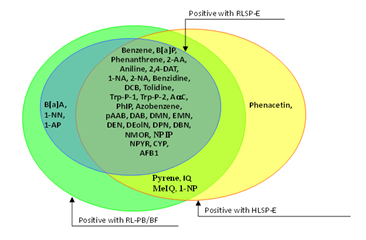
Figure 8 Pro-mutagens showed positive responses under human liver S9 from single donor and rat liver S9’s.
We used the comet assay to assess DNA damage as we compared the ability of S9 fractions from human and rat tissues to activate 37 pro-mutagens. A liver S9 prepared from rats pretreated with PB/BF or Aroclor 1254 to induce CYP is routinely used in in vitro genotoxicity tests.3 In spite of the higher activating ability of CYP-induced liver S9, only 4 pro-mutagens were activated by RL-PB/BF but not RLSP-E (pyrene, IQ, MeIQ, and 1-NP). These results suggest that CYP induction by PB/BF does not greatly affect whether pro-mutagens show a positive or negative response. It does not seem valid to compare the mutagenic magnitude of pro-mutagens activated by human versus rat organ S9 fractions, since there are great individual differences in CYP activity in human S9s. We believe that the importance is whether pro-mutagens show a positive or negative response is affected by species difference of S9, because routine use of human S9 in genotoxicity studies is difficult and we have been continuing to use data obtained from organisms other than humans to assess genotoxic risk in humans.
The level of CYP activity in human liver can be influenced by various life-style factors, including cigarette consumption.5 A possible explanation for the higher activity of CYP3A4 in the liver S9 from one human donor (HLS-014E, Table 3) is 10 years’ of enzyme induction by anti-asthma agents.5 Because of such great individual differences in CYP activity, liver S9 from one donor could not represent all human liver S9. It is also not likely that a pooled human liver S9 fraction, such as HLSP-E, which was prepared from a pool of 15 donors can represent all human liver S9. The use of such a pooled preparation, however, might minimize individual differences. For 5 pro-mutagens, discrepant results were obtained with two human liver S9’s, i.e., 2 chemicals (1-AP and 1-NN) were positive with HLS-014E but negative with HLSP-E, and 4 (pyrene, B[a]P, 2-AA, and MeIQ) were positive with HLSP-E but were negative with HLS-014E (Figure 5). So we believe that HLS-014E, which has exceptionally high CYP3A4 activity, activated a greater variety of pro-mutagens than HLSP-E. In contrast, human lung S9, with far lower CYP activity, activated fewer pro-mutagens than human liver S9. HLuSP-E did not activate benzene, DMN, EMN, and DEolN, which are activated by CYP2E1,13–15 that can be explained by the lack of CYP2E1 activity in HLuSP-E.
It is difficult to consider that even pooled liver S9 such as HLSP-E can represent human liver S9. To compare the activating ability of human and rat S9, therefore, we tentatively considered that a pro-mutagen was activated by human liver S9 if either HLSP-E or HLS-014E led to a positive response. Results of only 2 of the 37 studied pro-mutagens were discrepant, i.e., only one (phenacetin) was positive with human liver S9 but not with RL-PB/BF (Figure 3). Phenacetin has been shown to be borderline positive in a forward mutation test (6-thioguanine resistance) in V79 cells, only in the presence of hamster liver S9, and gave negative results in the presence of rat liver S9 or without any metabolic system,12 which coincide with our present result.
And only one (B[a]A) was positive with RL-PB/BF but not with human liver S9 (Figure 3). Both RL-PB/BF and human liver S9 activated pyrene, IQ, MeIQ, and 1-NP while RLSP-E did not (Figure 3). These results suggest that the activating ability of rat liver S9 qualitatively parallels that of human liver S9 and that the genotoxicity of some pro-mutagens is missed by the use of CYP un-induced rat liver S9. We do not deny that the use of human tissue samples in in vitro genotoxicity tests would permit more precise assessment of genotoxic risk for humans, but because of the unstable supply of human organs, the great individual differences in CYP activity (which make it difficult to obtain a standard human S9), and ethical matters, the use of rat liver S9 will continue. Our results show that activating ability of human liver S9 qualitatively parallels that of CYP-induced rat liver S9, even though the latter causes more extensive DNA damage. Thus, the currently used CYP-induced rat liver S9 fraction is suitable for the evaluation of the genotoxic risk for humans in spite of species difference in CYP isoforms.
The authors are grateful to Prof. Michael Morris (Hachinohe National College of Technology) for his English review.
The author declares no conflict of interest.

©2018 Kawaguchi, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.