MOJ
eISSN: 2379-6294


Review Article Volume 3 Issue 2
1Laboratory of Genotoxicity, Hachinohe National College of Technology, Japan
2Department of Pharmaceutical Health Care, Himeji Dokkyo University, Japan
Correspondence: Yu F Sasaki, Laboratory of Genotoxicity, Faculty of Chemical and Biological Engineering, Hachinohe National College of Technology, Tamonoki Uwanotai16-1, Hachinohe, Aomori 039-1192, Japan, Tel 8117 8277 296, Fax 8117 8277 296
Received: May 30, 2017 | Published: June 14, 2017
Citation: Sasaki YF, Kawaguchi S, Nakamura T. The power of the comet assay to detect low level genotoxicity and dna repair factors affecting its power. MOJ Toxicol. 2017;3(2):35–42. DOI: 10.15406/mojt.2017.03.00049
The comet assay is considered to be a rapid and sensitive procedure for quantitating DNA damage in mammalian cells. In this article, to interpret its outcomes adequately, its power to detect low level genotoxicity and factors affecting its power were reviewed. Although the development of initial lesions into alkali-labile sites and/or SSBs through repairing events is an important factor to support its detecting power, their repair reduce its power to detect SSBs as an initial DNA lesion. The acellular comet assay, in which slides with gel are prepared from untreated cells are exposed after lysis to test agents. Thus, detection of SSBs as initial lesions but not alkali-labile sites generated from DNA lesion such as alkylated bases and the power to detect low level SSBs as initial lesions is lower in the standard than in the acellular assay. The acellular comet assay would be practically used to detect SSBs as initial lesions. The inhibitors of re-synthesis andincisionsteps of the excision repair enhance and suppress comet-tail formation, respectively. Although the incision step of repairing process is necessary to support the sensitivity of the comet assay, its re-synthesis step reduces the power to detect low level genotoxicity. Although this assay can detect a wide variety of genotoxoic compounds and its power to detect genotoxicity were identical to that of the MN test, its power to detect low levels of genotoxic potential is inferior to that of the MN test. However, its power of to detect a low level of genotoxicity can be elevated to a level higher than that of the MN test by using DNA resynthesis inhibitors, such as araC and HU. In conclusion, the outcomes of comet assay should be interpretedappropriately in consideration of the action of DNA repair.
Keywords: comet assay, power to detect low level genotoxicity, re-synthesis step, incision step, excision repair, ssbs
AMN, α-amanitin; APC, aphidicolin; AraC, cytosine-1β-d-arabinofuanoside; BER, base excision repair; BLM, bleomycin; DDTTP, 2',3'-dideoxythymidine-5'-triphosphate; DDT, 2',3'-dideoxythymidine; EMS, ethyl methanesulfonate; ENU, ethyl nitrosourea; GGR, global genome repair; HU, hydroxyurea; LGD, lowest genotoxic dose; MMS, methyl methane sulfonate; NER, nucleotide excision repair; MNNG, n-methyl-n'-nitro-n-nitrosoguanidine; MNU, methyl nitrosourea; MNT, micronucleus test; 4NQO, 4-nitroquinoline 1 oxide; SSB, dna single strand break; TCR, transcription coupled dna repair; UV, ultra violet
The comet assay is a rapid procedure for quantitating DNA lesions in mammalian cells.1,2 During the last two decades, this assay has developed into a basic tool for use by investigators interested in research areas ranging from human and environmental biomonitoring to DNA repair processes to genetic toxicology.2 In this assay, cells are embedded in agarose, lysed in an alkaline buffer, and subjected to an electric current. Relaxed and broken DNA fragments stream further from the nucleus than intact DNA, so the extent of the DNA damage can be measured by the length of the stream (Figure 1). This method has several advantages: it is considered to be highly sensitive to DNA damage expressed as single strand breaks and alkali-labile sites, it uses data from individual cells, and few cells are required. Furthermore, DNA damage can be measured in the absence of mitotic activity, and some reports on the use of the comet assay as a method to detect genotoxicity in organs without mitotic activity have recently been published.3
The comet assay is considered to be a rapid and sensitive procedure for quantitating DNA damage in mammalian cells.1,2 Under alkaline (pH >13) conditions, the assay can detect single and double-stranded breaks, incomplete repair sites, alkali-labile sites, and also possibly both DNA–protein and DNA–DNA cross-links, in virtually any eukaryotic cell population that can be obtained as a single cell suspension (Figure 2). Base adducts and alkylated bases induced in cells develop into SSBs and/or alkali-labile sites that can be detected by the comet assay through repairing events.2 The importance is that this assay can detect SSBs as initial lesions and repair intermediates but not initial DNA lesions such as base adducts directly. Therefore, to interpret the outcomes of this assay adequately, it is important to understand how the repair system affects its detecting power and what it can detect. In this article, initial DNA lesions are defined as DNA lesions, such as SSBs, alkylated bases, base adducts, and pyramiding dimmers that are produced directly by the reaction between mutagens and DNA. SSBs that generated during the repairing process of initial lesions, such as alkylated bases, base adducts, and pyramiding dimmers, are considered to be secondary lesions.
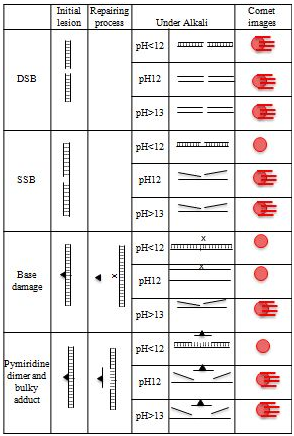
Figure 2 Mechanisms of comet-tail formation upon different kinds of initial lesion.▲, base lesion; x, AP-site.
The main purpose of this article is to review the power of the comet assay to detect low level genotoxicity and what factors can affect this assay.
The acellular comet assay is one modified version of the comet assay, in which comet slides hat are prepared from untreated cells are exposed after lysis to test agents and then processed according to the standard comet assay protocol.4 In the acellular comet assay, since lysed cells are exposed to test compounds, any biological events do not act to affect the formation and/or disappearance of SSBs (Figure 2). Kawaguchi et al.5 compared the power of standard and acellular comet assays to detect low level genotoxicity and reported as follows:
Mutagens |
Acellular |
Standard |
Ratio of Lgd at pH13 |
||
pH 12 |
pH >13 |
pH 12 |
pH >13 |
Acellula: Standard |
|
BLM |
0.0625 |
0.0625 |
2.5 |
2.5 |
1/40 |
MMS |
5 |
5 |
- |
40 |
1/8 |
EMS |
125 |
125 |
- |
1000 |
1/8 |
MNU |
12.5 |
12.5 |
- |
25 |
1/2 |
ENU |
31.25 |
31.25 |
- |
125 |
1/4 |
UV |
- |
- |
- |
10 |
|
Table 1 LGDs (mg/mL or J/m2) in acellular and standard acellular comet assays4
In this article, to compare the power to detect low level genotoxicity, the lowest genotoxic dose (LGD) is defined as the lowest dose at which each mutagen causes a positive response on each genotoxicity assay, i.e., a low LGD indicates a high power. BLM is well known to induce SSBs,6 which coincides well with the induction of comet-tails by BLM in the acellular and standard assays at pH 12. LGDs of BLM were 0.0625 and 1.25 µg/mL in the acellular and standard assays, respectively. This discrepancy is explained as follows; although the rejoining of BLM induced SSBs before comet preparation inhibits comet-tail formation, rejoining of BLM-induced SSBs does not occur in the acellular assay where any cellular function cannot act. The induction of SSBs by BLM at <1.25 µg/mL is considered to be too low to persist until comet preparation, being responsible for the negative and positive responses at <1.25 µg/mL in the standard and acellular assays, respectively.5 SSBs as initial DNA lesions that are not rejoinedareresponsible for comet tail formation. For SSB-inducers such as BLM, therefore, it is reasonable that comet-tails are produced in the acellular condition at pH 12 and pH >13 (Figure 3). On the other hand, in the standard assay, when the level of SSBs as initial DNA lesions is high enough to persist until comet preparation, comet-tail is expected to be observed.5 UV is well known not to induce SSBs as initial lesions but to induce pyramiding dimmers that are removed by the nucleotide excision repair and SSBs are formed during the repairing process of pyramiding dimmers. In the acellular assay, as shown in the case of UV, comet-tails cannot be formed for mutagens that do not produce SSBs as initial lesions.5 During the BER process of alkylated bases, AP-sites that can form SSBs at pH >12.6 are produced.7 For alkylating agents, therefore, a comet-tail is expected to be produced in the standard assay at pH >13 but not in the acellular assay (Figure 3).With contrast to this expectation, alkylating agents (MNU, ENU, MMS, and EMS) induce comet-tails in the acellular assay at pH 12 and pH >13 (Table 1).5 Since alkali-labile sites cannot be formed in the acellular condition where any cellular function cannot act, a comet-tail observed in the acellular condition at pH 12 and pH >13 reflects SSBs as initial lesions and it is discussed that those alkylating agents induce SSBs as initial lesions.5 Alkylating agents are classified into two types, unimolecular (SN-1) and bimolecular (SN-2) alkylating agents. Since LGDs of alkylating agents are lower in the acellular assay than in the standard assay, the power of the acellular assay to detect low level genotoxicity of alkylating agents is discussed to be higher than that of a standard assay.5 LGDs of SN-1 type agents (MNU and ENU) and SN-2 type agents (MMS and EMS) were >1/4-fold and 1/8-fold lower in the acellular assay than in the standard assay, respectively, from which it is suggested that SN-2 type agents produce SSBs more easily than SN-1 type agents.5 In general, SN-1 type alkylating agents produce primarily O-alkylated bases, whereas SN-2 type agents produce primarily N-alkylated bases.8 N-Alkylated bases such as 7-akyl pureness and 3-alkyl pureness are known to be liable to develop into AP sites that can form SSBs by β-elimination, which is discussed to coincide with the power of an acellular assay to detect low level genotoxicity is higher for SN-2 than SN-1 type agents.5 Since SSBs are generated from AP sites at pH >12.6,9 the superiority in electrophoresis condition of pH >13 is that not only SSBs as initial lesions and repair intermediates but also alkali-labile sites can be detected. Alkali-labile sites are generated from initial lesions other than SSBs by cellular function such as DNA repair.7 Although developing of initial lesions into alkali-labile sites through repairing events is a crucial factor to support the detecting power of the comet assay, repairing events reduce its power to detect initial DNA lesion such as SSBs. The power of the acellular comet assay to detect SSBs as initial lesions is higher than that of the standard assay. The acellular comet assay would be suitably used to detect SSBs as initial lesions.5
Although repair systems have widely divergent mechanisms, one step they all share is the re-synthesis of new DNA by DNA polymerases (Figure 4). APC which is an inhibitor of DNA polymerases10 can block the re-synthesis step of repairing process (Figure 5), thus causing the persistence of SSBs and enhancing comet-tail formation11-13 On the other hand, novobiocin interacting with the ATP as subunit of top isomerase II can inhibit the incision step and AMN interacting with RNA polymerase can inhibit the incision step of TCR, thus reducing repair-specific DNA cleavage and inhibiting comet-tail formation (Figure 4).13,14 Therefore, these DNA repair inhibitors could be used to suppress the repair steps (incision step and polymerization–ligation step) separately.15 Not only three replicative DNA polymerases, α, δ,and ε, but also one non-replicative polymerase, β have been identified as taking part in DNA repair pathways.10 The contributions of each polymerase vary depending on the specific damaging agent and the pathway of repair.11 Incision of UV-induced pyramiding dimmers creates a long gap of about 30 bp during NER, which must be re-synthesized by DNA polymerases δ/ε. On the other hand, BER creates a short gap, usually one nucleotide, but sometimes up to six.10 DNA polymerases b and δ/ε are involved in repairing gaps of one and 2-6 bp, respectively, during BER in mammalian cells.10 Although NER removes photoproducts upon UV exposure and bulky adducts upon exposure to a multitude of chemicals,10,16,17 BER removes altered bases (e.g., oxidized, methylated, and delaminated) that cause relatively minor disturbances to the helical DNA structure.18 Although DNA polymerases α, δ and ε are APC-sensitive polymerases, polymerase β is resistant to APC and sensitive to ddTTP.10 The effects of three kinds of DNA re-synthesis inhibitors on the formation of comet-tails are shown as follows; APC and the combination of araC and HU (araC/HU) enhanced comet-tail formation by UV, a UV-mimetic agent, 4NQO, and methylating agents, MMS and MNU and 2',3'-dideoxythymidine (ddT, a precursor of ddTTP) enhanced comet-tail formation by MMS and MNU, but not UV and 4NQO.19 During the incision step of repair processes, SSBs and/or alkali-labile sites are produced as intermediates, which can be visualized as comet-tails in the comet assay. Photoproducts produced by UV radiation and bulky adducts produced by a multitude of chemicals are removed by NER.17,18 On the other hand, MMS and MNU produce methylated bases in the DNA; the damage is repaired by the BER.20-24 DNA polymerase β is inhibited by ddTTP but not by APC and mammalian DNA polymerase β is the major DNA polymerase involved in the resynthesis step in BER.10 In human cells, the repair of 75% of BER-generated short gaps, usually one nucleotide, is estimated to be due to DNA polymerase β.25 BER-generated long gaps of 2 - 6 bp and NER-generated long gaps of 30 bp are closed by DNA polymerases δ/ε.10,25,26 It is discussed that ddT can enhance comet-tail induction by mutagens that produce DNA damage repaired by BER but not NER, and that APC and araC/HU can enhance comet-positive responses without mutagen specificity.21 Since APC inhibits DNA polymerase δ/ε but not b,10 the enhancement of a comet-tail formation by APC is considered to be due to the prevention of resealing by DNA polymerases δ/ε but not b19 The enhancement of4NQO- and UV-induced comet-tails by APC but not ddT is explained by the fact that DNA polymerase δ/ε but not β acts in NER.19 Although APC-resistant DNA polymerase b is the major DNA polymerase acting in BER, DNA polymerases δ/ε also acts in BER,10 which also explains the observation that APC enhanced comet-tail formation upon exposure to not only 4NQO and UV but also MMS and MNU.19 The inhibition of DNA re-synthesis by araC is due to either direct inhibition of DNA polymerase or indirect inhibition through ara CMP incorporation into a repaired region of DNA, rendering it unsuitable for further polymerase action.27 HU prevents DNA replication by selectively inhibiting rib nucleotide reductive11 Thus, the combination of araC and HU inhibits DNA resynthesis without polymerase specificity. Therefore, the enhancement of a comet-tail formation by araC/HU is believed be due to the sum of repair synthesis inhibition by DNA polymerasesb and δ/ε.20 Based on the observation that a comet-tail formation by exposure to mutagens that produce damaged bases removed by BER is enhanced by DDT, BER depending on DNA polymerase b can be distinguished from NER by the enhancement of a comet-positive response by DDT.19 Based on this discussion, it is studied how bulky alkylated bases can be removed by NER or BER: AMN reduced the comet-tail induction by n-alkyl Methanesulfonate having an n-alkyl group with >4 carbons and DDT enhanced it byn-alkyl Methanesulfonate having an n-alkyl group with <5 carbons, from each. It is discussed that NER acts to remove bases alkylated by n-alkyl groups with >4 carbons, BER acts to remove bases alkylated by n-alkyl groups with <5 carbons, and both NER and BER act to remove bases alkylated by n-alkyl groups with 4 and 5 carbons.15
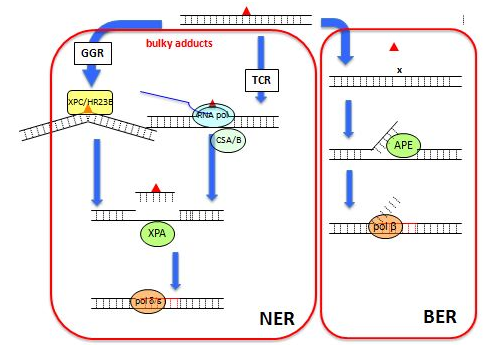
Figure 4 The process of NER and BER.
▲, base lesion; x, AP-site;AP Endo, AP endonuclease; GGR, global genomerepair; NER, nucleotide excision repair;BER: Base Excision Repair;pol b, DNA polymerase b; pol δ/ε, DNA polymerase δ/ε; RNA pol, RNA polymerase; TCR, transcription-coupled DNA repair.
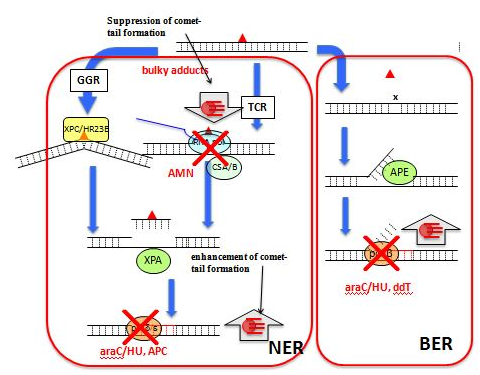
Figure 5 Expected effect of inhibition of NER and BERon comet-tail formation.
▲, base lesion; X, inhibition of an enzyme; x AP-site APE, AP endonuclease; GGR: Global Genome Repair; NER, nucleotide excision repair;BER: Base Excision Repair; pol b, DNA polymerase b; pol δ/ε, DNA polymerase δ/ε; RNA pol, RNA polymerase; TCR: Transcription Coupled DNA Repair.
One of the major DNA repair pathways is nucleotide excision repair(NER), which eliminates a variety of DNA lesions, including UVC-induced cyclobutane pyramiding dimmers (CPD) and (6-4) photoproducts (6-4PP), a wide variety of chemical bulky adducts, and certain types of DNAcross-links. There are two sub-pathways in NER.28 One is designated as TCR, which preferentially occurs in the transcribed strand of transcription ally active genes. The other is GGR, which occurs throughout the genome including the non-transcribed strand of active genes. The outcomes of the comet assay were discussed in three different NER-defective human cells (XPA cells derived from a xeroderma pigment sum (XP) group A patient, XPC cells derived from an XP group C patient, and CSA cells derived from a Cocaine syndrome (CS) group A patient)as follows: 1) comet-tails were formed by alkylating agents (MNU, ENU, BNU, and MMS) and BLM in the three NER-defective human cells, 2) UV induced comet-tails in XPC and CSA cells but not in XPA cells3) UV did not induce comet-tails in XPA not only in the absence of inhibitors of re-synthesis step of DNA repair but also in their presence, and4) the induction of comet-tails by UV was observed later in CSA than in NER-wild and XPC cells.29 Both NER sub-pathways are defective in XP groups A because of a defect in the XPA protein, and only GGR is defective in XP groups C because of a defect in the XPC protein,30 and only TCR sub-pathway is selectively deficient in CS because of a defect in the CS protein. Pyramiding dimmers and alkylated bases are induced by UV and alkylating agents, respectively. In the case of UV and alkylating agents, comet assay-detectable SSBs and/or alkali-labile sites are formed during the excision repair process.31 It is known that pyramiding dimmers and alkylated bases are repaired by NER and base excision repair (BER), respectively. XPA, XPC, and CSA cells are NER-mutant human cells with different deficiencies in NER, that is, both GGR and TCR sub-pathways are deficient in XPA cells, the GGR and TCR sub-pathways are selectively deficient in XPC and CSA cells, respectively. Because of the deficiency in both GGR and TCR, the incision of DNA strand in the vicinity of pyramiding dimmers does not occur in XPA cells, which results in the absence of the formation of comet tails.29 The incision of DNA strand occurs in XPC and CS cells by TCR and GGR, respectively, which can explain the formation of comet-tails in UV-irradiated XPC and CS cells.29 Therefore, it is discussed that UV can induce comet-tails, even if one of two sub-pathways of NER is impaired and that the incision step of NER is necessary to support the sensitivity of the comet assay.29
A tumor suppressor, the p53 protein, plays a key role as “guardian of the genome” by reducing the accumulation of mutations and inhibiting progression towards malignancy.32 It has been shown that p53 affects the pathway leading to mutation and the mutation frequency.33 The effect of p53 status on the outcomes of the comet assay was shown as follows (Table 2):
Mutagens |
Cell |
-araC/HU |
+araC/HU |
LGD Ratio |
BLM |
TK6 |
6.25 |
6.25 |
1 |
WTK1 |
6.25 |
6.25 |
||
MMS |
TK6 |
40 |
5 |
2 |
WTK1 |
20 |
5 |
||
EMS |
TK6 |
125 |
62.5 |
1 |
WTK1 |
125 |
62.5 |
||
MNU |
TK6 |
25 |
3.13 |
2 |
WTK1 |
12.5 |
3.13 |
||
ENU |
TK6 |
250 |
62.5 |
2 |
WTK1 |
125 |
62.5 |
||
UV |
TK6 |
20 |
1.25 |
8 |
WTK1 |
2.5 |
1.25 |
Table 2 LGDs (mg/mL or J/m2) with and without araC/HU in TK6 and WTK1 cells and LGDratio (TK6:WTK1) without araC/HU33
Here, TK6 and WTK1 cells are derivatives of human WIL-2 human lymphoblastoidcells and are p53+/- and P53-/-, respectively.34 It is discussed that SSB formation from UV-induced pyramiding dimmers through the incision step of nucleotide excision repair (followed by SSB rejoining) occurs earlier in TK6 cells than in WTK1 cells.33 SSB-formation that is followed by SSB-rejoining occurs earlier in cells with p53+/- than in those with p53-/- which would result in more rapid disappearance of SSB in the former than in the latter .33 Lower LGD in TK6 than in WTK1 cells without araC/HU is explained by the following possibility; the level of DNA lesions induced at low dose ranges of mutagens is low enough to be repaired before comet preparation in TK6 cells but not in WTK1 cells, i.e repair process proceeds more rapidly and SSB levels decreased more rapidly in TK6 cells with p53+/-than in WTK1 cells with p53-/-.33 Similar LGD in TK6 than in WTK1 cells in the presence of araC/HU is explained by the following possibility: since SSBs are not rejoined, even if the formation of SSBs occurs earlier in TK6 than in WTK1 cells, the accumulation of formed SSBs is similar in TK6 and WTK1 cells.33
When the sensitivity of a genotoxicity testing method is regarded as high, it means that it can detect a wide variety of compounds with unknown genotoxic potential and that the assay can detect a low level of genotoxic potential by known genotoxic compounds. The former is very important so as to avoid pseudo-negative results and the comet assay can detecta wide variety of genotoxic compounds both in vitro35,36 and in vivo.3,37-40 The micronucleus test (MN test) is a standard procedure that can detect structural chromosome aberrations derived from initial damage in the S phase and/or numerical chromosome aberrations due to an eugenic effects in the M phase.40,41 Pfau et al.42 conducted combined experiments with MLC5 cells including comet assay and MN test and showed that the genotoxicity of heterocyclic amines was observed at concentrations that were 2000-fold lower on an MN test than on a comet assay. However, van Goethe et al.43 showed that the LGDs of pure cobalt powder, a cobalt-containing alloy, and cobalt-tungsten carbide were lower on comet assay than on MN test. Hartmann et al.44 tested 36 pharmaceutical compounds with unknown genotoxic potential comparatively in the comet assay and the MN test using V79 Chinese hamster cells and reported that more compounds were positive in the MN test than in the comet assay. However, the exposure conditions used in some of those studies differed among different assays Pfau et al42 exposed cells for 30 min and 24 h in the comet assay and the MN test, respectively) and model mutagens with well characterized action mechanisms were not used, making it difficult to systematically compare the sensitivities of those assays including the comet assay. Kawaguchi et al.45 conducted combined experiments with TK6 cells including (standard) comet assay, acellular assay, and an MN test under identical exposure conditions. Results are summarized as follows (Table 3):
Mutagen |
Comet |
Comet/araC |
Acellular |
MN |
LGDratio |
UV |
20 |
2.5 |
- |
15 |
1.3 |
MNU |
25 |
3.125 |
6.25 |
12.5 |
2 |
ENU |
250 |
62.5 |
31.3 |
125 |
2 |
MMS |
40 |
20 |
10 |
20 |
2 |
EMS |
125 |
62.5 |
62.5 |
125 |
1 |
BLM |
12.5 |
6.25 |
0.0625 |
5 |
2.5 |
Table 3 LGDs (µg/mL or J/m2) for Comet assay, Comet assay/araC, acellular assay and MN test inTK6 and LGD ratio (Comet: MN)42
Based on these results, the detecting power is discussed as follows:
A comet assay can detect SSBs as an initial lesion and repair intermediates but not detect initial DNA lesions, such as base adducts, directly. Therefore, DNA repair events greatly affect its power. Although the comet assay has been considered to be a rapid and sensitive procedure for quantitating DNA damage in mammalian cells, its power to detect a low level of genotoxicity is not strong, so that it was considered conventionally and is rather inferior toan MN test. The power of a comet assay to detect a low level of genotoxicity is lower than that of an MN test. Its low power to detect a low level of genotoxicity is due to the disappearance of SSBs by repair process before slide preparation. Therefore, the canceling of the re-synthesis step of excision repair can elevate its power. On the contrary, the incision step of excision repair is necessary to support its power. In conclusion, although comet assay is a basic tool for use by investigators interested in research areas ranging from human and environmental biomonitoring to DNA repair processes to genetic toxicology, its outcomes should be interpretedappropriately in consideration of the action of DNA repair. For example, even UV cannot induce comet tails when the incision step of excision repair does not act, from which UV cannot be interpreted not to induce DNA damage in cells defective in the incision step of excision repair.
The authors are grateful to Prof. Michael Morris (Hachinohe National College of Technology) for his English review.
The author declares no conflict of interest.

©2017 Sasaki, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.