MOJ
eISSN: 2374-6920


Research Article Volume 7 Issue 2
1School of Biotechnology and Bioinformatics, D Y Patil University, India
2KIIT School of Biotechnology, KIIT University, India
Correspondence: Pritam Kumar Panda, KIIT School of Biotechnology, KIIT University, Bhubaneswar, India
Received: December 12, 2017 | Published: April 5, 2018
Citation: Shah N, Rao H, Verma SK, et al. Structure-based drug designing methodology to elucidate the binding affinities of phyto-ligands of Annona muricata against oncogenic PirH2. MOJ Proteomics Bioinform. 2018;7(2):113 ? 120. DOI: 10.15406/mojpb.2018.07.00220
The incidence of lung cancer is amassed in the current era due to sophisticated vicissitude living that serves as a pre-eminent cause of mortality. PirH2, an oncogenic protein encoded by RCHY1 gene has ubiquitin-protein ligase activity has been experimentally determined to interact with p53 tumor suppressor protein that has the ability to promote apoptosis. The oncogenic protein contributes directly to malignant tumor development as interaction with p53 promotes proteasomal degradation of p53. As a result, the loss of function of p53 due to DNA damage and abnormal signaling leads to malignant tumor(s). Thus regulation of PirH2 function is necessary to provide stability of p53 in response to several abnormal activities. Many experimental approaches have proven the fact that the interaction of p53 with PirH2 decreases the level of expression of p53 and repress its function. The in-silico work performed here focuses on finding a potential drug that could help in curing lung cancer and multiple other cancers as the drug targets cancer cells overexpressing PirH2. This involves homology modeling of PirH2 receptor, combinatorial library preparation of phytochemicals derived from Annona muricata (Graviola Plant), molecular docking and virtual screening studies of phytochemicals against the receptor to identify potential inhibitors that can serve to regulate the p53 activity by suppressing the expression of PirH2.
Keywords: PirH2, Graviola, I-GemDock, Autodock Vina, Virtual screening, PRODRG
Malignant neoplasm or cancer involves abnormal cell growth and by inchmeal it starts to proliferate over other parts of the body (http://www.who.int/mediacentre/factsheets/fs297/en/; http://www.cancer.gov/about-cancer/what-is-cancer). One of the latest worldwide incidences and mortality studies of 27 major cancers reveals lung cancer being the pre-eminent cause of cancer death (1.6 million) when compared to deaths caused by any other types of cancer (http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx). With no absolute prevention and definitive cure, finding a treatment for cancer has become a prime concern (http://www.who.int/mediacentre/factsheets/fs297/en/). This research paper focuses on finding a potential drug that could help in curing lung cancer and multiple other cancers as the drug targets cancer cells overexpressing PirH2; a protein discovered belonging to the family of ubiquitin protein ligase (http://www.yourgenome.org/facts/is-cancer-a-genetic-disease). Lung cancer is one of the major genetic diseases causing death1 and it is the most commonly diagnosed cancer in the world. Lung cancer can be broadly divided into three types: Non-Small cell Lung cancer, Small cell lung cancer and Lung Carcinoid tumor involving carcinomas. These tumors originate from epithelia of the trachea, bronchi or lungs, most common of them being squamous cell carcinoma, adenocarcinoma, and small cell carcinoma. Amongst the three types mentioned, the small cell lung cancer (SCLC) is known to be most malignant. Cancer cells are very sensitive to radiation and chemotherapy, chemotherapy being an essential component of the treatment of all patients with SCLC2 thus lung cancer bears emphasis in the following research.
A cell is a highly complex entity which carries out a function depending on the signaling it receives. When the cell undergoes a cellular damage like DNA-damage and is not able to repair itself, the cell is programmed to undergo apoptosis. But, a cancerous cell mislays its ability to signal itself to either repair DNA or perform apoptosis and starts rapidly proliferation, and if not treated promptly, it may also spread to other parts of the body. In cancer cells, apoptosis is not carried out because the apoptotic proteins are inactivated.3 Cancer cells are known to up-regulate multiple inhibitors of apoptosis proteins thus proliferating at a rapid rate.
Out of various apoptotic proteins, p53 is observed to be having reduced function of apoptosis in cancerous cells as PirH2 gets over-expressed.3,4 ‘PirH2’ is a protein of ubiquitin–proteasome pathway playing a crucial role in maintaining cellular protein homeostasis. It is known to ubiquitinate p53 protein responsible for apoptosis, and hence the protein gets degraded. Protein ubiquitination is catalyzed by a highly regulated enzymatic cascade, including ubiquitin-activating enzyme and ubiquitin conjugating enzyme E1s and E2s, along with ubiquitin ligase E3s, E3s primarily determine the substrate specificity and E3 ubiquitin ligases is compartmentalize into three major groups, RING (real interesting new gene) finger domain and HECT (the homologous to E6-AP carboxyl terminus) containing E3s along with U-box proteins.
Ubiquitin–proteasome pathway plays a crucial role in maintaining cellular protein homeostasis; involved in many cellular processes, such as cell proliferation, differentiation, DNA repair, apoptosis, and metabolism.5 Some of the inhibitors which follow ubiquitin-proteasome pathways like Mdm2 and PirH2 (p53 inducible E3 ligase with a RING-H2 domain) were observed to be up-regulated in multiple cancers.5,6 Mdm2 and PirH2 are both E3 (ubiquitination) ligases having a common target - p53 protein.7 As a result, in the case of any aberrant activities of E3 ubiquitin ligases is strongly correlated with the pathogenesis of various human diseases, including cancer.5
PirH2, being an E3 ligase has multiple substrates along with p53– c-Myc, p27Kip1, p27, p73, PolH (DNA polymerase η) and all of these are known to be responsible for either repair and/or apoptosis of the cell. c-Myc is a protein involved in growth control, differentiation and apoptosis. Its overexpression sensitized cells to apoptosis. p27(Kip1) is a cell cycle regulatory protein that interacts with cyclin-CDK2 and CDK4 which inhibits cell cycle progression at G1 and causes apoptosis.4 p73 is a protein related to p53 tumor protein structurally and is a tumor suppressor involved in cell cycle regulation and induction of apoptosis. There are various isoforms of this protein found in different tissues in an organism. Among the various DNA polymerases, DNA Pol η encoded by POLH gene and is involved in DNA repair by translesion synthesis and is activated only when the cell is under stress and DNA is damaged.6,7 CHK2 (Checkpoint Kinase 2) is a protein that serves as a checkpoint effector for signaling of dsDNA breaks as well as activation of cell cycle checkpoints. PirH2 interacts with CHK2 and mediates the proteasomal degradation.
All of these proteins are directly or indirectly involved with apoptosis of the cell. These proteins are targeted by E3 ligase-PirH2 and ubiquitylated for their degradation paving the way for the cell to survive. PirH2 is expressed in normal cells at normal concentrations, but in the case of cancerous cells, where apoptosis must occur, this protein is found to be up-regulated. Also, it has been studied for its action in tumorigenesis by suppressing p53 (tumor suppressor protein) and blocking it has been postulated as a new technique for developing anticancer drugs.3,8 Targeting PirH2 will not only plan to normalize the quantity of p53 but other apoptosis proteins as well, serving as a very advantageous target.
Certain fruits possess compounds – phytochemicals that help in reducing cancer and are thus recommended by medical physicians to cancerous patients.9 Annona muricata – Graviola (Soursoup) is one of the recommended fruit possessing anti-cancerous, anti-viral, and anti-inflammatory properties and is thus a reason for the study.10 Graviola is known to be associated with multiple cancers primarily lung cancer.11,12 A list of phytochemical compounds of Graviola and their structural analogs was obtained and their in-silico based interaction study was done with PirH2 receptor. PirH2 expression is up-regulated in multiple cancers like lung, prostate and head and neck and thus the drug interacting with PirH2 could also be used not just for lung cancers, but multiple cancers which overexpress Pirh2.3
In this paper, we are foreseeing the anti-cancer activity of phytochemicals in Graviola fruit extracts and their structural analogs against PirH2 by performing in-silico based binding studies as shown in the workflow (Figure1). The receptor PirH2 was docked with multiple phytochemicals that were obtained from Annona muricata. The drug “Muricatetrocin B” has shown potential to interact with PirH2 receptor with the highest stability in comparison to other compounds and thus can act as a potential drug inactivating or adversely affecting PirH2 receptor which results in reduced activity of the E3 ligase protein thus not causing anti-apoptosis leading to the natural death of cancerous cells.
This chemoinformatic study pivots on finding a compound, which would downregulate the ubiquitination activity of the Pirh2; hindering its process of poly-ubiquitination of apoptotic protein. Curtailed ubiquitination of proteins would help proteins perform apoptosis-like they would in normal cells.
The substrate/receptor
As depicted in Figure 1, the protein “PirH2” was identified as the target protein for the analysis. The E3 ubiquitin ligase PirH2 is responsible for proteasomal degradation of major target proteins like p53, p73 and also affects the stability of the translesion DNA polymerase Pol H thus contributing to the regulation of cell cycle progression. Thus, the protein was used for docking.
As per the literature review, the human protein PirH2 consist of 3 subunits i.e. RING-H2 domain, N-terminal, and C-terminal; NMR structures of the individual subunits were obtained from ‘RCSB PDB’ (http://www.rcsb.org/) and the PDB IDs for the same are 2JRJ, 2K2C, and 2K2D. The atomic coordinates for all the subunits were obtained using ‘solution NMR’. The three different subunits of the native protein were combined to form a stable protein using Hex 8.0.0.13
The protein PirH2 was selected because of its ubiquitin ligase activity causing proteasomal degradation of proteins responsible for cell survival and is overexpressed in the case of cancer, especially lung cancer. PirH2 (p53 inducible E3 ligase with a RING-H2 domain) has ubiquitin ligase activity and is regulated by p53 gene and functions irrespective of the activity of Mdm2. It is a 261 amino-acid protein and acts an apoptosis factor. The protein has three domains – N-Terminal domain (Amino acid 1-137), the RING domain (amino acid 138-189) and the C-terminal domain (amino acid 190-261). It has an unusually high concentration of cysteine (11%) and histidine (8%). This medium-sized protein also has nine zinc residues as shown in Figure 2.6
Prediction of Active Site
CastP14 server was used to predict the active site of the protein PirH2 and Accelrys Discovery Studio v4.515 for predicting its active site for molecular docking purpose. As blind docking was performed, the x, y, z Cartesian coordinates were chosen so as to cover the major core of the compound which was later used to prepare Grid Box in Auto Dock Vina for molecular docking.
Compound library preparation
List of 81 phytochemicals was obtained from16 and structure for the same was downloaded from PubChem and HMDB Database. Certain structures were obtained in 2D format, which was converted to 3D using ChemAxon - Marvin Sketch (https://www.chemaxon.com/products/marvin/marvinsketch/). These compounds were optimized geometrically by Arguslab (www.arguslab.com). These compounds were virtually screened and their analogs of compounds from (Table 1) were obtained with >90% similarity from ZINC database (http://zinc.docking.org/) in PDB format and their properties were taken into consideration. These original compounds and analogs were further virtually screened. Geometrical optimization was performed on all the ligands and analogs using ChemAxon (Marvin Sketch).
Compound ID |
Compound Name |
Energy(kcal/mol) |
PubChem11028548 |
cis-Annomontacin |
-116.6 |
HMDB41428 |
Muricatalin |
-113.7 |
HMDB31168 |
Cohibin A |
-109.4 |
PubChem10698767 |
cis Annonacin |
-107.4 |
HMDB35900 |
Muricatenol |
-106.7 |
PubChem393572 |
Muricatetrocin B |
-106.5 |
PubChem44566987 |
Annocatalin |
-106.2 |
HMDB40918 |
Corepoxylone |
-105.9 |
Table 1 Best selected compounds by virtual screening of 81 phytochemicals with protein PirH2 and their respective energies.
IDs |
NAME |
Vina Docking Score (kcal/mol) |
Interacting H-atoms of protein molecule with ligand |
Length (A0) |
CASTp analysis |
HMDB31168 |
Cohibin A |
-5.3 |
THR43, |
2.344, |
Allosteric Site 4 |
HMDB35900 |
Muricatenol |
-4.8 |
TYR86, |
3.306, |
Catalytic site and Allosteric site 4 |
HMDB40918 |
Corepoxylone |
-5.5 |
ASP88, |
2.088, |
Allosteric site 4 |
HMDB41428 |
Muricatalin |
-6.6 |
ASN65, |
2.242, |
Allosteric Site 4 |
PubChem393472 |
Muricatetrocin B |
-6.8 |
CYS20, |
2.252, |
Allosteric Site 4 |
PubChem10698767 |
cis Annonacin |
-5.4 |
CYS20, |
2.347, |
Allosteric site 4 |
PubChem11028548 |
cis-annomontacin |
-5.0 |
ARG54, |
2.098, |
Allosteric site 2, |
PubChem44566987 |
annocatalin |
-6.2 |
ASN11, |
2.896, |
Catalytic site |
Analogs |
|||||
ZINC ID 42806832 |
cisannomontacin3-0 |
-6.6 |
GLN14, |
2.176, |
Catalytic site and allosteric site 4 |
ZINC ID 42806832 |
muricatetrocinb-1-1 |
-7.0 |
ILE89, |
2.083, |
Catalytic site |
ZINC ID 42806832 |
muricatalin2-0 |
-6.7 |
GLN72, |
2.504, |
Catalytic site and Allosteric site 4 |
ZINC ID 42806821 |
cisannonacin1-1. |
-6.4 |
GLN14, |
2.939, |
Catalytic site |
ZINC ID 42806827 |
muricatetrocinb-3-0 |
-5.8 |
|
|
- |
ZINC ID 42806821 |
annocatalin1-0 |
-6.6 |
ASP88 |
2.015 |
Allosteric site 4 |
ZINC ID 42806827 |
cisannonacin2-1 |
-6.8 |
|
|
- |
ZINC ID 42806839 |
muricatetrocinb-2-0 |
-6.6 |
ASN65, |
2.248, |
Allosteric site 4 |
ZINC ID 42806821 |
cohibina-10-0 |
-6.6 |
ASP88 |
2.195, |
Allosteric site 4 |
ZINC ID 42806827 |
muricatenol-3-1 |
-6.8 |
LEU39, |
2.604, |
Catalytic site |
ZINC ID 42806821 |
muricatenol-4-0.pdb |
-6.7 |
|
|
- |
ZINC ID 42806827 |
cohibina-9-0.pdb |
-6.2 |
ASN104, |
2.215, |
Catalytic site and Allosteric site 7 |
ZINC ID 42806832 |
muricatenol-2-0.pdb |
-6.8 |
LEU21, |
2.194, |
Catalytic site and Allosteric site 4 |
ZINC ID 42806821 |
muricatalin4-1.pdb |
-6.8 |
GLY106, |
2.078, |
Catalytic site and Allosteric site 4 |
Table 2 Molecular docking score of PirH2 along with the best of original compounds and selected conformers. The table also includes all the details of binding affinities and the site of interaction
Analog preparation
A total of 44 analogs were obtained using ZINC database with >90% similarity. These analogs were subjected to PRODRG server to acquire the most stable conformation. The small molecule topology generator PRODRG takes Cartesian coordinates and converts into a wide range of topologies suitable for various analyses like GROMACS, docking, protein-protein docking, a compound with polar hydrogen, and compounds with hydrogen molecules merged, etc. The top eight compounds selected by virtual screening and the 44 conformers were used for further studies. The PRODRG server will take small molecules, which are phytochemicals in this case and generate an energy minimized, coordinates in a wide range of topologies for use with various software.17
Pharmacophore-based virtual vcreening
CADD method was used to recognize and find potential leads by virtually screening these ligands by iGemDock v2.118with the receptor PirH2, which resulted in a selection of a total of 14 conformers (Table 2). These were virtually screened and the following compounds were known to show a high binding affinity for the receptor.
Rigid docking
22 ligands were docked with PirH2 receptor by Hex 8.0.0 with standard parameters and their energy was taken into consideration (Table 3). Hex was used to perform rigid docking to estimate the ligand binding affinities and to study their interaction with PirH2. The complex was saved and the interaction between the ligand-receptor was observed using Accelrys Discovery Studio Visualizer v4.5.
IDs |
NAME |
Etotal (kcal/mol) |
Amino acid of PirH2 interacting with the ligand |
HMDB31168 |
Cohibin A |
-345.41 |
LYS A:58 |
HMDB35900 |
Muricatenol |
-402.14 |
GLU A:47 |
HMDB40918 |
Corepoxylone |
-340.95 |
ILE A:89 |
HMDB41428 |
Muricatalin |
-351.24 |
ILE A:89 ASP A:88 GLN A:72 ASN A:65 THR A:43 ASN A:41 HIS A:70 TYR A:86 |
PubC393472 |
Muricatetrocin B |
-321.04 |
- |
PubC10698767 |
cis Annonacin |
-364.04 |
ARG A:72 |
PubC11028548 |
cis-annomontacin |
-374.92 |
GLN A:51 |
PubC44566987 |
annocatalin |
-425.17 |
ASN A:11 GLY A:37 LEU A:39 |
42806832 |
cisannomontacin3-0 |
-346.85 |
- |
42806832 |
muricatetrocinb-1-1 |
-346.85 |
- |
42806832 |
muricatalin2-0 |
-346.85 |
- |
42806821 |
cisannonacin1-1 |
-374.28 |
- |
42806827 |
muricatetrocinb-3-0 |
-372.59 |
- |
42806821 |
annocatalin1-0 |
-374.28 |
- |
42806827 |
cisannonacin2-1 |
-372.59 |
- |
42806839 |
muricatetrocinb-2-0 |
-356.32 |
HIS A:38 |
42806821 |
cohibina-10-0 |
-374.28 |
- |
42806827 |
muricatenol-3-1 |
-372.59 |
- |
42806821 |
muricatenol-4-0 |
-374.28 |
|
42806827 |
cohibina-9-0 |
-372.59 |
- |
42806832 |
muricatenol-2-0 |
-346.85 |
- |
42806821 |
muricatalin4-1 |
-374.28 |
- |
Table 3 Rigid Docking performed by Hex 8.0.0 showing their binding affinity energy and the various amino acids that are interacting
Molinspiration
It is important to understand the molecular properties and predict the bioactivity of the compounds for a drug to be compatible with the human in-vivo environment. These properties are important enough to be studied as they predict the possible stability inside the body. Lipinski’s rule of five11 was also taken into consideration. Eight original compounds and 14 conformers of analogs were uploaded on the Molinspiration server to predict its structural properties and their bioactivity. Refer (Table 4) for the same. The data estimated by the software can be used to predict the potential and capacity of these compounds to be drugs, and can be helpful to compare the compound’s biochemical properties.
IDs |
HMDB 31168 |
HMDB 35900 |
HMDB 40918 |
HMDB 41428 |
PubChem 393472 |
PubChem 10698767 |
PubChem 11028548 |
PubChem 44566987 |
logP |
9.76 |
9.34 |
9.29 |
8.12 |
2.74 |
7.28 |
8.26 |
7.28 |
TPSA |
69.92 |
110.37 |
74.75 |
139.84 |
116.45 |
116.45 |
116.45 |
116.45 |
Molecular Weight |
554.94 |
614.992 |
566.91 |
616.92 |
472.66 |
598.91 |
626.96 |
116.45 |
violations |
2 |
2 |
2 |
3 |
0 |
2 |
2 |
2 |
rotb |
29 |
31 |
27 |
26 |
17 |
26 |
28 |
26 |
Volume |
622.54 |
672.23 |
610.89 |
644.87 |
479.75 |
630.96 |
664.57 |
630.96 |
GPCR ligand |
0.18 |
0.09 |
0.24 |
0.13 |
0.24 |
0.12 |
0.03 |
0.14 |
Ion Channel Modulator |
-0.2 |
-0.48 |
-0.21 |
-0.53 |
-0.17 |
-0.49 |
-0.69 |
-0.52 |
Kinase Inhibitor |
-0.11 |
-0.27 |
0.01 |
-0.19 |
0.08 |
-0.23 |
-0.38 |
-0.22 |
Nuclear Receptor Ligand |
0.04 |
-0.14 |
0.14 |
-0.14 |
0.1 |
-0.12 |
-0.28 |
-0.09 |
Protease Inhibitor |
0.31 |
0.3 |
0.46 |
0.38 |
0.29 |
0.21 |
0.16 |
0.22 |
Enzyme Inhibitor |
0.18 |
0.01 |
0.32 |
0.2 |
0.48 |
0.18 |
0.02 |
0.21 |
Table 4 Molecular properties and predicted bioactivity by Molinspiration
Molecular docking
It is essential for designing a drug to understand the interactions between the protein molecule – PirH2 and the ligand molecules, the molecules highly interacting with the protein would be able to actually. Thus, molecular docking plays a crucial role in understanding the interactions predicted between the ligand and the receptor. Molecular docking studies were carried out using AutoDock Vina 4.2.19 Blind Docking was performed with the Grid Box cube of 40 and spacing of 1 with the x, y, and z centers as 5.964, 2.304, and 3.298. Preparation of protein was carried out using these parameters and by adding Kollman charges20 and merging non-polar hydrogen atoms. The ligands were prepared using AutoDock by applying Gasteiger charges19 and merging non-polar hydrogen atoms. 14 poses were generated for each ligand based on the best-suited orientation of the molecule. Table 3 represents the molecular docking scores of PirH2 with compounds of original confirmation and their “Zinc” analogs serving a comparative analysis of their stability in the form of energy and also the hydrogen-bonds that are formed with their respective atoms in the amino acids. The amino-acids were later correlated to their respective cavity in the protein with the help of CastP.
Autodock Vina docking results showed better affinity to compounds presented in Table 3. The active site for the PirH2 receptor was predicted using ‘CastP’ server, which yielded various cavities in the structure and the largest cavity was presumed to be the active site. Muricatetrocin B showed intermolecular H-bonding with RCHY1 at CYS20, GLU22, THR43, ALA71 with an energy of -6.8kcal/mol. Out of the whole compound library, conformer of an analog of Muricatetrocin B, i.e. Muricatetrocin B-1 (ZINC ID42806832) showed the strongest interaction towards PirH2 receptor with an energy of -7.0kcal/mol by the formation of intermolecular H-bonds with ILE89, CYS87, ASN60 of RCHY1. Although, the docking studies cannot comment on the inhibitory or stimulatory nature of the ligand’s interaction with the protein. The 44 phytochemicals initially obtained were scaled down to eight after the virtual screening based on the energy calculated by iGemDock. These eight compounds (Table 1) were predicted to be compounds with the potential to interact with PirH2. Their conformation and interaction can be visualized. Their respective structural analogs were retrieved from ‘ZINC database’ and the best conformer of these 44 analogs was generated using ‘PRODRG’ server. These conformers were also virtually screened using ‘iGemDock v2.1;18 14 out of those 44 portrayed good interaction and these 14 structural analogs and the original eight compounds obtained from initial virtual screening studies were further used for docking studies (Figure 1) (Figure 3).
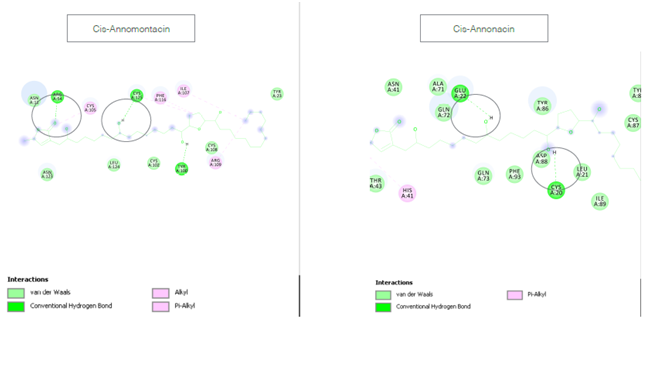
Figure 3 Binding interactions of ligands with receptor PirH2 as shown using 2D plot in Discovery Studio 2.0.
Molecular docking using autodock vina v4.2
Molecular docking was performed using AutoDock Vina to study binding affinities of protein-ligand interaction. Grid box was prepared for the selected receptor and zinc ions were introduced in the virtual grid dimension created for docking interaction; zinc ions are fundamentally required for structural stability and integrity of PirH2 receptor.
The docking score was calculated by AutoDock Vina and Muricatetrocin B and Muricatalin showed to bind with PirH2 with the energy of -6.8 kcal/mol and -6.6 kcal/mol respectively. The docked structures showing less binding energy are shown as follow in the (Table 2). The 2D plot of binding interactions was shown using Discovery Studio v4.5 as shown in Supplementary Table 1).
Cancer has become prevalent nowadays, as the number of people diagnosed with it is increasing every decade and it is further expected to rise from 14 million in 2012 to 22 within the next 2 decades (http://www.who.int/mediacentre/factsheets/fs297/en/).The survival rate of the cancer patients hasn’t commensurated well enough with the existing available chemotherapeutic treatments against various types of cancer which have been proven to be clinically successful. Since there isn’t any definite rationale finding which truly illuminates us with the mechanism of cancer development and metastasis. This quest for finding the absolute preventive and as well as a cure, prevailed the very happening of this in-silico based analysis, which suggests the use of certain phytochemicals present in ‘Graviola’ fruit extracts and their predicted structural analog as potential compounds which can be used in developing chemotherapeutic drugs against cancer cells which overexpress Pirh2. This study provides implication toward the possible usage of these phytochemicals as prospective drug candidates. Although, wet lab studies of these compounds on cancer cell lines is indispensable, in order to validate the biological activity of these molecules which would further pave the way to use these compounds as a remedy to cure various types of cancer.
None.
Author declare there is no conflict of interest.

©2018 Shah, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.