MOJ
eISSN: 2374-6920


Research Article Volume 1 Issue 4
1School of Biosciences and Technology, VIT Vellore, India
2Department of Biotechnology and Bioinformatics, Bishop Heber College, India
3Applied Mathematics Department, M S University, India
4Department of Bioinformatics, St. Aloysius College, India
5CompCYC - A Virtual Informatics Research Organization, India
Correspondence: Jitendra Kumar Gupta, Department of Bioinformatics, St. Aloysius College, Mangalore, Karnataka, India, Tel +918123089626
Received: July 01, 2014 | Published: August 8, 2014
Citation: Pathak A, Madar IH, Raithatha K, et al. In-silico identification of potential inhibitors against AChE using cheminformatics approach. MOJ Proteomics Bioinform. 2014;1(4):96-100. DOI: 10.15406/mojpb.2014.01.00022
Acetylcholinesterase (AChE) is highly viable target in Alzheimer’s disease (AD) as cholinergic deficit is consistent and early finding in AD. From previous studies, it has been proved that the Cholinesterase inhibitors (ChE-Is) are the only class of drugs approved by the Food and Drug Administration (FDA) for treatment of AD. Comparison of inhibitory activity of 15 different ChE-Is class of drugs was performed using cheminformatics and molecular docking approach against AChE. This approach helps to identify the binding affinity, mode of interaction and to understand the selectivity of drug molecule for the treatment of Alzheimer’s disease. This article suggests the best market available ChE-Is for AD.
Keywords: acetyl cholinesterase (ACheE), alzheimer’s disease, acetyl cholinesterase inhibitors (AChE-I), huperziaserrata
ACheE, acetyl cholinesterase; AChE-I, acetyl cholinesterase inhibitors; FDA, food and drug administration; AD, alzheimer’s disease; APP, amyloid precursor protein; YASARA, yet another scientific artificial reality application; GA, genetic algorithm
Alzheimer’s disease (AD) is a neurodegenerative disorder characterized by brain shrinkage results from a grouping of genetic, lifestyle and environmental factors that affect the brain over time. It is one of the most rigorous health problems in old age people.1 Approximate 5% of people with the disease have early onset of Alzheimer’s (also known as younger-onset), which repeatedly appears in the age of 40s or 50s. AD patient has decline in cognitive dysfunction, primarily memory loss and in the later stages of the disease, depression, agitation, language deficits, and psychosis and mood disturbances.2,3 AD patients have shown progressive damage to brain cells by forming Amyloid Plaques, neurofibrillary tangles and subsequent loss in cholinergic neurons.4 Acetylcholine is found in cholinergic neurons located in most of the areas of brain and spinal cord. When cholinergic neurons release acetylcholine it diffuses across the gap to receptors on a neighboring neuron, to make cell-cell signaling. The enzyme acetylcholinesterase (AChE) hydrolyses the chemical acetylcholine as soon it is released in the system.
Though the damage and destruction of cholinergic cells cannot be controlled, but the performance of brain can be temporarily boosted by maintaining the level of acetylcholine as high as possible.5 These drugs inhibit the hydrolysis of acetylcholine by acetyl cholinesterase and helps in its proper transmission. Thus AD treatment has specifically dependent on inhibition of acetylcholinestarases.6
Molecular docking helps in predicting the intermolecular interactions of drugs/ligands to specific active binding site of the protein.7,8 The modifications in protein functions can be reveled by visualizing protein-ligand complexes formed after docking, which is very important part of drug development.8 The energy values of all 15 protein-drug complexes were compared in this article.
Preparation of 3D structures
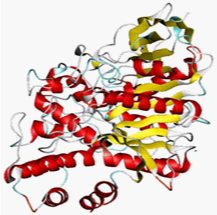
3D structure of AChE was retrieved from Protein Data Bank PDB_ID:1B41 (Figure 1). SMILES strings of all 15 drug molecules were obtained from Pub Chem and Drug Bank database.9 The molecular structures were remodeled using Chem Sketch and CORINA. Open Babel10 was used for file format conversion.
Determination of active site
The structural binding pockets and active sites in the protein were determined by literature search.
Q-SiteFinder11 was used to validate the catalytic residues in the protein, which retrieves the binding sites by calculating interaction energy between simple imaginary Vander Waals probes on the surface of protein structure.
Protein structure minimization
YASARA program was used to perform structure optimization. YASARA stands for “Yet Another Scientific Artificial Reality Application”. Stability of protein structure increases with the decrease in energy of the protein structure. It is important to assess the energy of structure before performing docking. The energy of the structure before and after minimization was 204284.4KJ/ Mol, −286744.6KJ/ Mol respectively, which proves the minimized structure, is more stable.
Molecular docking and interaction study
Autodock,12 a tool to perform molecular docking of protein with virtually screened ligands to compute the binding energy of docked complexes. The 3D structure of protein was used in PDBQT format. Each structure has been transformed and refined using Kollman and Gasteiger charges along with the polar hydrogen molecules. Number of rotatable bonds was decided and Non-polar hydrogen atoms were fused. The grid points have been generated using an Autogrid program in Autodock tools (Grid Size - x=y=z=60) and spacing of 0.375Ao was set.
Docking simulations have been performed using Genetic Algorithm (GA). By default 10 runs were performed for each docking experiment that was set to halt after a maximum of 250000 energy calculations. The default parameter for best conformation search, population size=150, translational steps=0.2Å, quaternion steps=5 and torsion steps=5 were applied.
Pathway linker
Pathway Linker13 was used for categorization and visualization of the first neighbor network interacting to target protein. It explores the signaling pathway of proteins in the sub network and offers links to related online services (Figure 2).
LAMB1: Laminin subunit beta-1 is a protein in humans encoded by the LAMB1 gene.
COL4A1: Collagen alpha-1(IV) chain is a protein in humans encoded by the COL4A1 gene.
APP: Amyloid precursor protein (APP) is an integral membrane protein expressed in many tissues and concentrated in the synapses of neurons.
LAMA1: Laminin subunit alpha-1 is a protein that in humans is encoded by the LAMA1 gene.
HAND1: Heart- and neural crest derivatives-expressed protein 1 is encoded by the HAND1 gene.
COLQ: Acetylcholinesterase collagenic tail peptide is also known as AChE Q subunit, acetylcholinesterase-associated collagen, or ColQ encoded by the COLQ gene, is the collagen-tail subunit of acetylcholinesterase.
LGTN: Ligatin is a protein in humans, encoded by the LGTN gene.
The experiment shows that Ser-203, His-447 and Glu-334 falls on the catalytic binding site of AChE, evaluated using Q-SiteFinder.9 It was also found that catalytic site residues: Ser-203 and His-447 are present in the first predicted site of volume 652Å and Glu-334 is present in the fourth predicted site of volume 185Å. These catalytic site residues are shown in three dimensional structure of AChE (Figure 3).
Molecular docking of 15 drugs has been performed by using Autodock Tools4.2.12 The Binding energy, Torsional Free Energy, Final Total Internal Energy, vdW + Hbond + deSolv Energy, Electrostatic Energy was calculated in kcal/Mol, whereas Inhibition constant is in micro molar (uM). The interaction complex of AChE-Ambenonium complex (Figure 4c) shows positive ΔG value, indicating binding is not stable. The binding affinity in Ache-Huperzine-A (Figure 4e) complex is best compared to other complexes. The energy and Ki values have been summarized in Table 1. The molecules with minimum energy and which had shown binding with the active site of Acetylcholinesterase has been selected. They are Huperzine A,14 Galantamine, Tesofensine, Tolserine, and Donepezil.15–17 The interaction energy of AChE complexes with Huperzine A and complexes with Galantamine showed better values compared to other three complexes. So based on the Lipinski Rule18 and the Partition coefficient concepts Donepezil, Tolserine, Tesofensine drug molecules were discarded as they have higher partition coefficient more than 4, which is not good for drug absorption.
S. no |
Drug name |
BE (ΔG) |
RMSD (A°) |
Ki (uM) |
Final intermolecular energy |
IE |
TE |
|
VdW-Hb-dE |
EE |
|||||||
1 |
Physostigmine |
-8.16 |
209.51 |
1.05 |
-7.63 |
-1.12 |
-0.07 |
0.60 |
2 |
Donepezil |
-8.25 |
210.99 |
0.889 |
-9.56 |
-0.49 |
1.78 |
1.79 |
3 |
Huperzine A |
-10.93 |
208.05 |
0.009 |
-9.17 |
-2.06 |
0.15 |
0.30 |
4 |
Rivastigmine |
-7.27 |
214.561 |
4.68 |
-7.72 |
-1.05 |
-0.22 |
1.49 |
5 |
Tacrine |
-7.66 |
210.623 |
2.43 |
-7.78 |
-0.18 |
0.06 |
0.30 |
6 |
Ambenonium |
9.22 |
205.954 |
-- |
6.42 |
-1.67 |
15.2 |
4.47 |
7 |
Galantamine |
-9.22 |
207.45 |
0.17 |
-8.67 |
-1.14 |
-0.06 |
0.60 |
8 |
Chlorpyrifos |
-6.63 |
209.92 |
13.71 |
-8.43 |
0.00 |
-0.34 |
1.79 |
9 |
Pyridostigmine |
-5.51 |
223.437 |
92.12 |
-5.99 |
-0.12 |
-0.19 |
0.60 |
10 |
Edrophonium |
-6.45 |
211.418 |
18.69 |
-6.21 |
-1.13 |
-0.14 |
0.89 |
11 |
Metrifonate |
-4.45 |
208.976 |
544.31 |
-5.76 |
-0.19 |
0.77 |
1.49 |
12 |
ladostigil |
-8.82 |
210.254 |
0.343 |
-9.42 |
-0.89 |
-0.31 |
1.49 |
13 |
Phenserine |
-9.67 |
209.95 |
0.081 |
-9.93 |
-0.64 |
-0.13 |
0.89 |
14 |
Tolserine |
-8.53 |
209.114 |
0.562 |
-8.77 |
-0.65 |
-0.26 |
0.89 |
15 |
Tesofensine |
-8.91 |
207.798 |
0.294 |
-8.82 |
-1.28 |
17.9 |
1.19 |
Table 1 Docking analysis of AChE with selected drug molecules
TE, torsional free energy in (kcal/Mol); IE, final total internal energy in (kcal/Mol); BE, free binding energy in (kcal/Mol); vdW-Hb-dE, vdW + Hbond + desolve energy in (kcal/Mol); EE, electrostatic energy (kcal/Mol); Ki, inhibition constant in (uM); RMSD, root mean square deviation in Å
When compared with AChE-Galantamine complex, AChE-Huperzine-A complex has shown lower binding energy; also has better interactions. Huperzine A, is a Lycopodium alkaloid, a dietary supplement extracted from the Chinese club moss Huperziaserrata. It inhibits acetylcholinesterase and acts as an NMDA receptor antagonist, which helps in boosting the memory by raising the brain neurotransmitter. It was found to have an antioxidant and neuroprotective properties which is helpful for AD patients. Huperzine A works well for young as well as aged patients (Figure 4a-4o).
Based on molecular docking results it is revealed that Huperzine A is the best drug for AD when compared with other 14 drugs available in market. Many others properties of a drugs are also to be taken into consideration before claiming the above. This research work can be a support to various on-going in-vitro experiments.
None.
The author declares no conflict of interest.

©2014 Pathak, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.