MOJ
eISSN: 2374-6920


Review Article Volume 7 Issue 1
1Department of Pathology, Sylhet Agricultural University, Bangladesh
2College of Veterinary Medicine & Bio-Safety Research Institute, Chonbuk National University, South Korea
3Department of Medicine, Sylhet Agricultural University, Bangladesh
4Department of Pathology, Bangladesh Agricultural University, Bangladesh
Correspondence: Md. Masudur Rahman, Department of Pathology, Faculty of Veterinary, Animal and Biomedical Sciences, Sylhet Agricultural University, Sylhet 3100, Bangladesh, Tel +88-0821-761002(201), Fax +88-0821-761980
Received: December 20, 2017 | Published: January 19, 2018
Citation: Runa NS, Husna A, Yesmin S, et al. Ebola virus: an updated review on immunity and vaccine. MOJ Proteomics Bioinform. 2018;7(1):1-7. DOI: 10.15406/mojpb.2018.07.00205
Ebola virus (EBOV) is one of the most dangerous viruses in the world which causes fatal hemorrhagic fever syndrome both in humans and nonhuman primates. Major innate immunity mechanisms against EBOV are associated with the production of interferon’s that are inhibited by viral proteins. Activation of host NK cells was recognized as a leading immune function responsible for recovery of EBOV infected people. Uncontrolled cell infection by EBOV leads to an impairment of immunity. EBOV proteins interaction with host cellular proteins disrupt type I and type II interferon responses, RNAi antiviral responses, antigen presentation, T-cell-dependent B cell responses, humoral immunity, and cell-mediated immunity. These multifaceted approaches to evasion and suppression of innate and adaptive immune responses in their target hosts lead to the severe immune deregulation and "cytokine storm" that is characteristic of fatal EBOV infection. Long-term control of viral outbreaks requires the use of vaccines to impart acquired resistance and ensuing protection. Development of a safe and efficacious vaccine against EBOV has proven elusive so far, but various inventive strategies are now being employed to counteract the threat of outbreaks caused by EBOV and related filoviruses. This review highlights the host immune responses to EBOV infection, its ability to subvert host immunity and discuss recent advances in prevention of EBOV infection by vaccination.
Keywords: ebola virus, immunity, vaccine, nucleic acid detection, cytokine storm
EBOV, ebola virus; EVD, ebola virus disease; NAT, nucleic acid detection; VLPs, virus like particles
Ebola virus (EBOV) derived its name from the Ebola River in Democratic Republic of Congo (DRC) (formerly Zaire) where the first Ebola virus disease (EVD) outbreak was observed in 1976.1 EBOV is enveloped, non-segmented, negative-stranded RNA virus containing filamentous virion which belongs to the family Filoviridae. EVD is caused by filamentous EBOV associated with mortality rates between 25 and 90%.2 The clinical symptoms of this disease are non-specific and flu-like, such as high fever, headache and myalgia at the early stage.3 The symptoms also included diarrhea and vomiting when the large outbreak occurs in West Africa (2014 to present).4 Immune cell disorders are associated at the late stage of this disease such as, diffuse intravascular coagulopathy caused by coagulation and neutrophilia disorders.5–7 Humans may acquire the disease by close contact with biological fluids of the infected animals or patients. During the acute phase of illness, EBOV has been detected in different body fluids including breast milk, saliva, semen, stool, sweat, tears and urine.8–12 EBOV has been isolated from urine (9days) and from aqueous humor (9weeks) after the virus was cleared from plasma.13
It has been observed that there is a correlation between the survival following EBOV infection and the ability of the host to mount an early and robust interferon response. In cultured human liver cells, the virus has blocked the innate immune response which helps in preventing the expression of interferon-stimulated genes.14,15 The experiments in animal models, including mice16,17 and guinea pigs18 have shown that blocking of interferon signaling is essential for the pathogenesis of the disease. The emergence of EBOV should be considered under the dual perspective of a large field of emerging viruses and considerable complexity and diversity among EBOV themselves. Unexpected viral outbreaks affecting humans over the past century include Yellow fever, Chikungunya, Dengue, West Nile Virus, MERS-CoV, Enterovirus 68, Enterovirus 71, Zikavirus, Japanese encephalitis, Hantavirus, Lassa, Marburg, Rift Valley, Crimean-Congo fever and Nipah. In relation to other viruses, Ebola virus is large and long and almost visible in an optical microscope but it has small genome coding for 7 genes. Fruit bats are the main reservoir of Ebola virus but rodents, and perhaps other mammals may also play a role.2
The pathogen involved in EVD outbreaks in West Africa during 2014–2015 was Zaire Ebola virus identified by RT-PCR analysis and sequencing. The variant involved in this outbreak has been named Makona, by the name of a river running through the area between Guinea, Liberia and Sierra Leone where the outbreak was first occurred. Pending further investigations, the outbreak was probably a zoonotic event, which was transmitted to humans via an index case in Guéckédou district, Guinea;19 then human to human transmission ensued.
The clinical symptom of the current EVD outbreak in West Africa, in general, similar to that described in prior EBOV epidemics. The incubation period for person-to-person transmission of EVD typically ranges from 8 to 11days, but cases were reported with incubation periods as short as 2 and as long as 21days. Shorter incubation periods may be observed following direct inoculation of virus through injection with contaminated needles.4,20 Patients often present to health care providers within 1week after the symptom onset.21,22 In the early clinical sign of EVD, patients manifest signs and symptoms that mimic common tropical illnesses (e.g., dengue, malaria, typhoid fever and other viral infections23–25). The onset of the disease includes nonspecific clinical signs such as fever, headache, arthralgia, myalgia and back pain. Progressive gastrointestinal (GI) symptoms often arise within 3 to 5days of symptom onset.26,27 GI manifestations include abdominal pain, anorexia, nausea, vomiting and diarrhea, which lead to marked electrolyte imbalance, intravascular volume depletion and shock. Conjunctival infection, rash, hiccups, respiratory and neurologic findings have been also reported. Bleeding is a late clinical sign that occurs only in less than 20% of patients with EVD.4 Rapid and reliable diagnosis of EVD is needed for appropriate and effective patient management, hospital or health center infection prevention and control, and optimization of use of healthcare resources.28 EBOV-specific laboratory tests were used to diagnose the specific suspected cases which applied to detect the EBOV genome (e.g., RT-PCR) or measurement of the EBOV antigen or specific antibodies.29 In the past 10months, the West Africa EVD outbreak has stimulated the development of new diagnostic tests, including rapid antigen detection tests and nucleic acid detection (NAT) tests such as loop-mediated isothermal amplification (LAMP) assays.30,31
Clinical trials for two vaccine candidates, the chimpanzee-adenovirus ChAd3-Zaire Ebola virus (ChAd3-ZEBOV) vaccine and the recombinant vesicular stomatitis virus-Zaire Ebola virus (rVSV-ZEBOV) vaccine has also been accelerated with the support of the World Health Organization. It has been noted that these therapeutics and vaccines are among the few that made it so quickly from the laboratory to the field. WHO and its international partners have invested a considerable amount of effort to fast-track clinical trials for these two vaccines. These efforts were rewarded six months after the outbreak was officially declared, when Phase I clinical trials for the ChAd3-ZEBOV vaccine, developed in the United States, began in the United Kingdom and the US (September 2014), as well as in Mali and Switzerland (October 2014). The rVSV-ZEBOV vaccine, developed in Canada, also began trials in the US (October 2014) and Gabon, Germany, and Switzerland (November 2014), as well as Kenya and Canada (December 2014). Both the ChAd3-ZEBOV and the rVSV-ZEBOV vaccines began Phase III clinical trials at the beginning of 2015. Initial results from an open-label, cluster randomized ring vaccination trial with rVSV-ZEBOV conducted in Guinea showed a vaccine efficacy of 100%.32 This review will focus on the evolution of filoviruses, with an in-depth look into the Ebola virus immunity and vaccine development.
Ebola virus is non-segmented, enveloped, single stranded, negative-sense RNA virus. About 300~1500 nm long EBOV contains 18,959 to 18,961 nucleotides. EBOV belongs to filovirus family and all of the viruses of this family are associated with characteristic filamentous particles which give the name of this virus family.33 These viruses contain genomes which are approximately 19 kb and contain eight proteins, VP35, VP40, VP30, VP24, the nucleoprotein (NP), glycoprotein (GP), soluble GP and L (polymerase) proteins.34 Except the glycoprotein gene, all of the genes are monocistronic, which encode for one structural protein. The RNA genome present in the inner ribonucleoprotein complex of virion particles that is encapsulated by the nucleoprotein. The structure of EBOV is shown in Figure 1. The viral genome associates with RNA-dependent RNA polymerase, VP35 and VP30 to the functional transcriptase–replicase complex.35 The proteins of the ribonucleoprotein complex are interferon antagonist which has another important function such as the role of VP35.34 The function of VP40 is to serve as the matrix protein and remotes particle formation.36 Another protein VP24 that is associated with the membrane which interposes with interferon signaling.37 There is a most important difference of EBOV from other Mononegavirales that they have ability to produce soluble glycoprotein, which is the main product of the GP gene, and infected cells secrete large quantities of virus.38,39
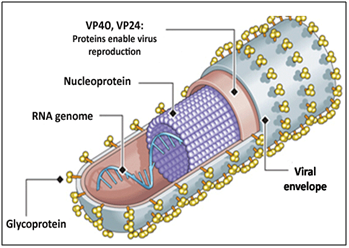
Figure 1 Structure of Ebola virus. VP40, VP24 are viral proteins that induce viral reproduction; single stranded RNA genome present in the inner ribonucleoprotein complex of virion particles that is encapsulated by the nucleoprotein; viral envelop contain large number of glycoprotein. (Source: http://www.creative-diagnostics.com/tag-ebola-virus-antigens-58.htm)
Impaired innate immune responses during filovirus infection
The innate immune system is the main basis for recognizing and cornerstone to eliminate effectively viral infections; rapid recognition of the microbe and subsequent activation of the host immune function. The entry pathway of Ebola virus in host cell is shown in Figure 2. Innate immune response is a key to develop effective adaptive immunity which invades pathogens. In both activation of innate immunity and initiation of adaptive immunity antigen-presenting cells are mandatory including macrophages, monocytes and dendritic cells (DCs). Antigen-presenting cells cause immune responses by the help of cytokines and chemokines; antigen presentation; interactions with B cell, T cell, and NK cells; and direct cytotoxic activity against target cells.40,41 In filovirus infections, many key notes support a critical role for innate immunity.
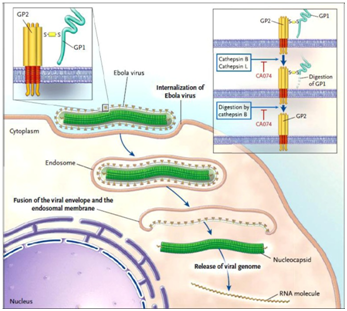
Figure 2 Entry pathway of Ebola Virus into host cell.64
Upon binding to cell-surface receptors, Ebola gets internalized in endosome. Within endosome, endosomal proteases: cathepsin B and cathepsin L, slash the viral GP1 protein into N-terminal fragment and then cathepsin B digests it further into only GP2. GP2 aids in the fusion of viral envelope and endosomal membrane, releasing viral genome into the cytoplasm. Upon release the proteolysis of GP1 is prevented by CA074 (inhibitor) and therefore infection advances.
First of all, survivors with filovirus infection have an early and short-lived associated with chemokines that is rise in serum which is indicative of innate immune system induction.42–46 Recent studies of EBOV-infected individuals provides an evidence that non survivors develop very high levels (5–1000X) of pro-inflammatory cytokines (IL-1β, IL-8, IL-15,IL-1RA, IL-6 and IL-16) and chemokines (MIP-1α, MIP-1β, MCP-1, eotaxin, MIF, and IP-10 GRO-α) that start to rise shortly after onset of disease and it continues within 2-3days before death until the last sampling.47 May be the primary mediators of this inflammatory response is virus infected monocytes and macrophages and the resulting secretion of cytokines and chemokines which enhance the permeability of endothelial layer and induces shock.48,49 One of the most important things is that both survivors and non-survivors do not have difference of important regulators of adaptive immunity such as IFN-α, IFN-γ, IL-12, IL-17, or TNF in their serum levels.47 The filoviruses also encode interferon antagonists VP24 and VP35, which block interferon production and inhibit downstream interferon signaling.50
Filoviruses evade the immune system by preventing the maturation of DC, which is important mediator for innate and adaptive immunity51,52 and it also encodes the multiple viral proteins that help in evasion of interferon responses50 By blocking of DC maturation EBOV infection inhibits the activation of lymphocytes and then eliminates those subsets that are capable of causing an effective response to the virus. EBOV smoothly infects and rapidly starts to replicate in antigen-presenting cells such as macrophages, monocytes, and DCs to produce large amounts of progeny virus.49,51–55 In macaques monkey after infection, infected DCs may disseminate virus throughout the body after trafficking from the infection site and that’s why the number of circulating HLA-DR+ cells increase 10X in blood. In spite of increased numbers in the circulation, infected DCs fail to become activated or mature and, therefore, after infection they are unable to induce appropriate B cell, NK cell and T cell responses.51,52 Monocytes and macrophages activity may also be failed, but in response to filovirus infection, they are an important issue in filovirology.49,51,52 For a certainly, in fatal cases of filovirus infection the downstream effects of antigen-presenting cell dysfunction are immeasurable with a marked lack of adaptive immunity noted.
Maintenance and activation of natural killer (NK) cells seems to be a vital to protect against lethal filovirus infection.56–58 however, in human and NHPs (non human primates) during filovirus infection NK cells and other lymphocytes are decreased.6,44,59,60 Although a Fas/FasL interaction is likely involved, NK and T cells are invisible in periphery that is due to apoptosis by a yet unidentified mechanism.47,59,61,62 Study in nonhuman primates, was observed a relationship between NK cells with the rapid protection against filovirus hemorrhagic fever that was induced by rVSV vaccine as a post exposure treatment.57 In rodents injected with VLPs 1–3days can also induce rapid and potent innate immune responses. Injected VLPs recruited nearly twice the number of NK cells in both the mediastinal lymph node and spleen compared to animals receiving PBS alone.58 This finding indicates that administration of VLP induces NK cell proliferation in mice lymphoid tissues with VLP-pretreatment lacking functional NK cells63,64 while, using antiasialoGM1 antibodies in mice with depleted NK cells did not protect from EBOV infection, unlike VLP injected wild-type C57Bl/6mice.
Moreover, native mice against EBOV infection protected by the adoptive transfer of NK cells which are stimulated in association with VLPs. Activation of the protective innate immunity requires performing, but not IFN- γ.58 On the surface of NK and T cells, KIRs are expressed and helped in activation of NK cells.47 Together, these data indicate that the cytolytic “killer” functions of NK cells are an important mechanism in protecting from EBOV infection and also play an important role for survival of NK cells from filovirus infection. Innate immunity combined with a lag becomes failing and causes inactivation of adaptive immune responses that induces an uncontrolled dissemination of filovirus infection.51,52,57
Role of T cells: In EBOV survivors, the early and apparently inflammatory response that is regulated followed by a detectable T cell response. It suggests that the activation of cytotoxic T cells (CTL).42,45,59 But in EBOV survivors, combined with detectable T cell responses an early and transient IgM is followed quickly by increasing levels of EBOV-specific IgG.42,59,65 Immune responses develop rapidly in the EBOV survivors which help to indicate that a swift induction of the appropriate immune responses in humans can result in survival from filovirus infection.59
After EBOV infection in mice both CD4+ and CD8+ T cells are depleted in the blood and spleen caused by apoptosis, similarly to humans and nonhuman primates. However, subsequent studies have shown that despite their massive loss in numbers, T cell function is maintained in the remaining cells. In controlling the disease and the damage done by the infection, T cells are sufficient to transfer to newly infected animals but at the late phase of infection the number of functional CD8+ T cells which are generated is likely too low to control high viral titers. The infection could be too severe to overcome but at that time the adaptive immune system can mount a response.66,67
EBOV specific laboratory tests are used to confirm the diagnosis of suspected cases which detect the EBOV genome (e.g., RT-PCR) or measure the EBOV antigen or specific antibodies.29 Last 10months ago, EVD outbreak in the West Africa, stimulated the development of new diagnostic tests that are rapid antigen detection tests and nucleic acid detection [NAT] tests such as loop-mediated isothermal amplification [LAMP] assays.30,31
Enzyme-Linked Immunosorbent Assay (ELISA)
Antigen detection: For EBOV detection, Antigen detection methods (e.g., enzyme-linked immunosorbent assay (ELISA)) were used as the gold standard method prior to 2000.65 ELISA has a relatively high sensitivity (93%) in the acute phase of EVD but during disease progression the antigen levels become decline, rendering lower sensitivity for antigen detection 1–2weeks following symptom onset.60,68 Many antigen detection tests are currently under evaluation and in the near future it may be extended to complement RT-PCR testing.30 RT-PCR is used in testing instead of ELISA which permits more rapid detection of Ag and can now be extended in mobile (portable) testing platforms in outbreak settings.69
Antibody detection: ELISA test is used to detect IgM antibodies against EBOV. This test is performed in the first week after the onset of symptoms. It is associated with a peak of IgM levels that occur in the 2ndweek of illness.25,65,68 From 1 to 6months of illness, IgM antibodies are found at variable rates.68 During EVD diagnosis many data showed that serology can be highly specific for this but it may be less sensitive in the intensive care unit setting. Therefore, in the diagnosis and management of critically ill EVD patients, antibody testing may be less useful.26 After the IgM antibodies, IgG antibodies appear soon which may persist for years68 but before the development of IgG antibody response, a substantial number of EVD patients have died.25
Reverse transcription polymerase chain reaction (RT-PCR)
For EVD diagnosis, RT-PCR is referred as the gold standard test. It is considered because of their high sensitivity and specificity. This test is used to detect the Ebola viral genome. This is generally performed by international mobile teams deployed in institutions such as the European Mobile Laboratory or CDC. To detect EBOV nucleic acid this test is used as highly sensitive nucleic acid amplification test.70 The sensitivity and specificity of this test are approximately 100% and 97%, respectively.43 If the test is performed within the first 3days of illness, molecular assays cannot detect the viral genome. For this it may leads to false negative results. So therefore, it should be repeated in subsequent samples.26 Proper sampling, collection, storage or transportation must be done to minimize the false-negative results and to avoid cross-contamination, a proper RT-PCR technique have to be implemented.2,26 Since data suggest high viremia associated with unfavorable outcomes and death, quantitative RT-PCR has been developed which is used to monitor the viral load.21,22 For rapid diagnosis within 10-30minutes, portable PCR techniques have been developed. These techniques do not require laboratory infrastructure.71 Portable PCR techniques perform more important role in disease surveillance and control associated with Ebola outbreaks and other infectious diseases.72
For infectious diseases in the human population, vaccination is considered one of the primary control strategies; however, there was no vaccine approved for use by the beginning of the last Ebola outbreak (2014-2016).The most important point to generate EBOV vaccines consist of classical formulations of inactivated virus with different adjuvant, such as Ribi adjuvant or lipid A-containing liposome.73–75 In 1980s Ebola vaccine development was started. After initiating, many vaccine candidates have been tested in rodents and NHPs, including inactivated virus, DNA vaccines, virus-like particles (VLPs) and also vaccines based on recombinant viral vectors.76 To test the efficacy of vaccine candidates many animal models have been developed, including guinea pigs, mice and other NHPs. But the Rodent models do not always helps in prediction of vaccine efficacy in NHPs. NHPs mostly rhesus and cynomolgus macaque is infected by non-adapted strains and causes best mimic disease progression in human. Therefore they are considered as the ‘‘reference’’ animal model for vaccine studies.73 There have many differences found in the EBOV antigen for vaccine development where the EBOV surface GP has been developed as main antigen and also the VLPs-based vaccine included the NP and the VP40 matrix protein also used. Replication deficient recombinant EBOV lacking the gene which encodes VP30 and it is an essential transcription factor for viral replication,77 and recently it has been shown to protect the NHPs against EBOV infection.78
Multiple approaches have been tested and demonstrated the protection against lethality in non-human primate models which may be partial to complete including treatment of the coagulopathic diathesis by use of nematode anticoagulant protein c2 (a potent inhibitor of tissue factor which initiated blood coagulation)79 or recombinant human-activated protein C (has a broad spectrum of coagulation-modulating activity), immune modulators such as human-activated protein C,80 therapeutic vaccines56,81,82 and gene-specific antivirals. Antiviral such as siRNA and phosphodiamidate morpholino oligomers directly inhibit the replication of EBOV. This antiviral appears to be lead candidates for the treatment of highly lethal filovirus infections which based on highly successful efficacy studies in NHPs.83–86 However in all cases, lower peak viral loads are linked with more promising outcomes in humans and NHPs.59,85
DNA vaccine
DNA vaccines are purified plasmid preparations. It contains one or more DNA sequences of the pathogen of interest which is elicited in the recipient on delivery (Figure 3). A DNA vaccine with ZEBOV GP expressed a protective immune response in mice87 and protected guinea pigs partially.88 A prime-boost vaccine strategy has developed. This strategy is made of four inoculations with DNA vectors which encoding ZEBOV GP and SEBOV GP and a boost with a recombinant adenovirus 5 vector expressing ZEBOV GP. This strategy generated cross-protective immunity in NHPs against a heterologous challenge with BEBOV.89 Currently, there have no licensed DNA vaccines for use in humans. However, after three injections of vaccine in a phase I clinical trial, a three-plasmid DNA vaccine encoding ZEBOV GP, SEBOV GP and NP expressed a cellular and humoral immune response in humans.90
Recombinant viral vector vaccines
Recombinant viral vector vaccines are consisting of vectors which are capable of replication or of replication-deficient vectors (Figure 3). The adenoviral vector is a most important replication-deficient vaccine vector which has a broad cellular tropism. It is used in a wide variety of gene therapy studies. Combination of DNA vaccine prime and adenovirus vector boost produced a vaccine strategy. It was able to protect NHPs from lethal BEBOV challenge.89 A single adenovirus vector with incorporating ZEBOV and SEBOV GP provided complete protection in NHPs against ZEBOV and SEBOV challenge. It demonstrates the potential of generating immunity against different EBOV species in NHPs.91 However, a lately completed phase I clinical trial substantiated that recombinant adenovirus serotype 5 expressing ZEBOV and SEBOV GP was safe and can be used in humans.92
Virus-like particle vaccines
VLPs are used in the vaccine production which is generated by the expression of one or more viral proteins and mimic the natural viral conformation. But it does not contain viral genetic material. EBOV VLPs were generated by simultaneously expressing GP, NP and VP40 of ZEBOV in 293T cell. These particles administered three times to NHPs combined with Ribi adjuvant to protect against homologous ZEBOV challenge.93 Recently another vaccine candidate has been investigated that is a replication-deficient ZEBOV which lack the VP30 gene. When two vaccines are administered with this virus, they protect mice and guinea pigs from lethal ZEBOV challenge.94
The review focuses on the Ebola virus infection, its immunity and vaccine development. Ebola virus infection leads to early and robust interferon-like responses that take place before the appearance of virus in the blood. Detectable T cell responses regulated an early and immediate response to the infection. Filovirus-specific CD8+ CTLs are necessary for viral control and clearance. The roles of T cells in protection are being assessed as multiple vaccine candidates are tested in NHP efficacy trials and, ultimately, in clinical trial volunteers. There is an urgent need for interventions to blunt the impact of EBOV on global health which has stimulated rapid progress in our understanding of EBOV biology, pathogenesis, and immunity. However, further investigation of immune responses in both natural filovirus infection and active immunzation in humans must be continued and expanded. For prevention and control of this lethal disease there is a prudent need for generating effective EBOV vaccines and increasing awareness among people.
This study was supported by a grant of the University Grants Commission (UGC) of Bangladesh (UGC Research Grant No, 5921) through Sylhet Agricultural University Research System (SAURES), Government of the People’s Republic of Bangladesh. The funder had no role in study design, data collection, and preparation of the manuscript or decision to publish.
The authors declare that no conflict of interest exists. No writing assistance was utilized in the production of this review article.

©2018 Runa, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.