MOJ
eISSN: 2374-6920


Research Article Volume 2 Issue 6
School of Life Sciences, Manipal University Dubai, UAE
Correspondence: Mohammad Zeeshan Ansari, School of Life Sciences, Manipal University Dubai, Dubai International Academic City, Dubai, United Arab Emirates, Tel +97144290736, Fax +97143694541
Received: October 25, 2015 | Published: November 20, 2015
Citation: Muhammad HM, Shabbir Z, Thogaru SR, et al. Computational approach towards targeting aggregate formation in synucleinopathies. MOJ Proteomics Bioinform. 2015;2(6):181-184. DOI: 10.15406/mojpb.2015.02.00066
Many neurodegenerative diseases such as Parkinson’s disease and Alzheimer’s disease are result of synucleinopathies, where neuronal cells show proteinaceous aggregates primarily formed of alpha-synuclein. Understanding the initial steps is crucial toward developing therapeutic strategies to combat these maladies. Using computational approaches, we attempted to understand the initial steps of oligomerization process of SNCA. Using predicted docked models of SNCA dimers, we identified important residues involved in the interaction of the assembly. Using combinatorial library of peptides and docking studies, we found several peptides that have better binding energies than wild type SNCA monomers. These findings will be valuable in guiding rational therapeutic interventions towards major neurodegenerative disorders involving synucleinopathies.
Keywords: synucleinopathies, parkinson’s disease, alzheimer’s disease, protein-protein interactions, docking, drug design
PD, parkinson’s disease; DLB, dementia with lewy bodies; MSA, multiple system atrophy; AD, alzheimer’s disease; SNCA, alpha-synuclein; SNCB, beta-synuclein (SNCB); SNCG, gamma-synuclein (SNCG); PIC, protein interactions calculator; PMV, python-based molecular viewer; VMD, visual molecular dynamics
Synucleinopathies is a diverse group of neurodegenerative diseases including Parkinson’s disease (PD), dementia with Lewy bodies (DLB), multiple system atrophy (MSA) and Alzheimer’s disease (AD) among others.1 A common characteristic of these synucleinopathies is presence of intracellular pathologic inclusions in certain brain cells that are primarily composed of α-synuclein. Based on the appearance of these proteinaceous aggregates in the affected cells, they are called with different names such as Lewy bodies and Lewy neurites in PD and DLB2 and glial cytoplasmic inclusions in MSA.3 Oligomerization of α-synuclein is the first step towards formation of these aggregates, which causes inflammation and ultimately leads to the neuronal cell death in these neurodegenerative disorders.4 A prion-like propagation of aggregates from affected cells to the nearby cells has also been proposed and therefore targeting the initial oligomerization steps remains one of the primary focus area for therapeutic strategies towards synucleinopathies.
Alpha-synuclein (SNCA) is one of the highly expressed cytosolic proteins in human brain. In regions like neocortex, hippocampus, substantia nigra, thalamus and cerebellum, its expression reaches as large as 1% of total cytosolic protein content.5 Despite more than two decades since its first identification in AD patients, the exact and complete functions of this 140 amino acid long protein is still elusive. However, the functional importance of SNCA in neurons and glials can be realized from these studies which showed that SNCA physically interacted with about 50 ligands and more than 30 proteins. Possible functions has been suggested in synaptic vesicle release and trafficking, fatty acid binding, physiological regulation of certain enzymes and transporters, and neuronal survival.6,7 SNCA is a 140 amino acid long protein encoded by SNCA gene on 4q21. In addition to this predominant form, alternate splicing produces at least two other shorter variants. The major form of SNCA can be distinguished structurally into three regions; amphipathic N-terminal (1-60), central hydrophobic (61-95) and acidic and proline-rich C-terminal (96-140) region.8 It belongs to class of natively unfolded proteins which are the most disordered subfamily member of intrinsically disordered proteins family. Environment, nevertheless, has a marked effect on its structure and SNCA transforms into a partially folded globular structure in acidic pH and high temperatures.9 On binding to lipid surfaces such as lipid droplets or lipid bilayer in vesicle membrane, SNCA adopted an elongated helical structure and SNCA monomers formed dimers and oligomers.10 In synucleinopathies, SNCA forms insoluble fibril aggregates which are multimers formed through such conformational changes and the central hydrophobic region plays and important role in protein aggregation.2 Interestingly, stabilization of monomers and non-fibrillogenic oligomers, on the other hand, have shown to have an inhibitory effect on the aggregate formation.11
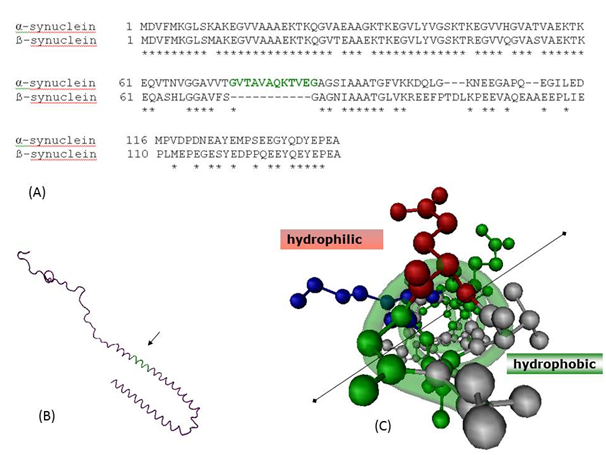
Two other members of synuclein family are β-synuclein (SNCB) and γ-synuclein (SNCG) located on 5q35 and 10q23.2, respectively. The predominant expressions of α- and β-synuclein expression overlap in the same regions of brain, while γ-synuclein is primarily expressed in peripheral nervous system and retina.12 Sequence comparisons reveal deletion of 11-mer region (73-83 of SNCA) in β-synuclein as shown in Figure 1.13 Despite having similar expression profile and homology, where SNCA is ubiquitously found in synucleinopathies, SNCB and SNCG are not found in Lewy bodies, the hallmark of PD. In this study, we have used computational tools to understand the role of this 11-mer peptide, its involvement in the dimerization of SNCA, and discuss ways to use this insight towards synucleinopathies therapeutics.
Sequence and structure analysis of SNCA
Sequence comparison of SNCA and SNCB were performed using BLAST web server at National Center for Biotechnology Information (NCBI).14 The NMR-structure of micelle-bound SNCA (PDBID 1XQ8) was obtained from Protein Data Bank.15,16 All visualization of protein structures were performed using Visual Molecular Dynamic (VMD).17
Protein-protein interactions during dimerization of SNCA
Two protein-docking experiments were performed: first between two SNCA molecules and second between an SNCA and the coordinates corresponding to the 11-mer peptide obtained from the SNCA. Web interface of protein docking software GRAMM-X from Vakser Lab was used.18 GRAMM-X and its web interface extend the original GRAMM Fast Fourier Transformation methodology by employing smoothed potentials, refinement stage, and knowledge-based scoring. The top-scoring models predict the interactions involving the 11-mer region. To explore the kind of interactions that stabilizes the assembly, we used Protein Interactions Calculator (PIC) server.19
Combinatorial library of 11-mer peptide
A library of 11-mer peptide was generated which contained various hydrophobic residues at those positions of 11-mer which were involved in the stabilization of SNCA dimer. Command-line program SCWRL 4 from Dunbrak Lab was obtained and used to generate the library.20 SCWRL is designed specifically for the task of prediction of side-chain conformations without changing the backbone coordinates of a protein. Though, in theory one can generate all possible N-mer peptides to generate a peptide library for evaluation of ligand-peptides that bind to the 11-mer region stronger than WT peptide, however, this will produce a huge number of candidate peptides (2011) and thus would be practically very difficult. In order to be within our capacity of analyses, only the residues found to be involved in interaction were computationally mutated to Leu, Ile or Val. The choices of these residues were based on the results from PIC server which confirmed that the interactions involved only hydrophobic interactions. We also avoided mutation to Phe, an amino acid with neutral aromatic side chains, as inclusion of phenyl group into any molecule drastically affects its lipophilicity and such molecules might not be very suitable as drug because of their ADME (adsorption, distribution, metabolism and excretion) properties.
Docking of the 11-mer combinatorial library with SNCA
To perform docking of each of the 11-mer peptide with SNCA protein, AutoDock 4.2 from The Scripps Research Institute was used.21 AutoDock along with the associated AutoDockTools were used to prepare each of the ligands and protein for docking. A 3D grid, which specifies the search space, was prepared around the 11-mer region of the SNCA and subsequent docking of the peptide from the combinatorial library was performed on this region of the protein. The docking results were viewed and analysed using PMV, a freely distributed Python-based molecular viewer.22
Sequence and structure analysis of SNCA
Figure 1a shows the pair-wise sequence alignment between SNCA and SNCB. The two sequences show significant similarity as expected, with E-value of 9E-43 and 70% similarity. The 11-mer region absent in SNCB is indicated in green colour in the sequence and structure of SNCA. This region adopts a helical structure upon membrane binding and the top view across the helix axis clearly shows the amphipathic nature with one side polar and other side majorly non-polar (Figure 1c). Since it is this region which is present in SNCA with respect to SNCB, we attempted to computationally explore this region which could be involved in fibrillation of SNCA in various synucleinopathies.
Protein-protein interactions during dimerization of SNCA
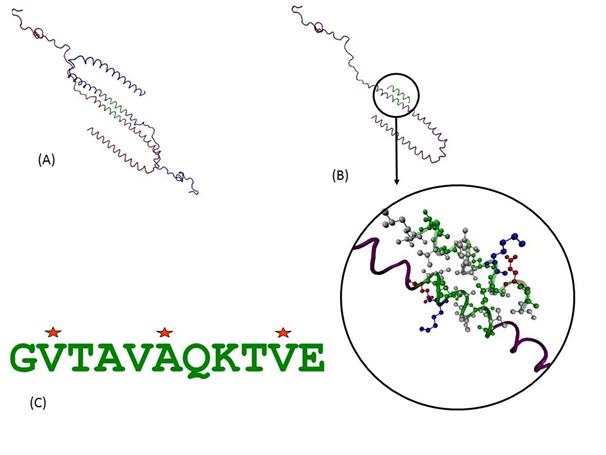
Protein-protein interactions can provide a wealth of information about function and behaviour of molecules. As there was no available structure for dimer or oligomer of SNCA, we used available state of the art computational prediction methods to address this crucial step. Detailed analyses of predicted protein-protein and protein-peptide docking interactions of the initial assemblies indicate that the interactions are purely hydrophobic in nature and are indeed predicted to be mediated through this 11-mer peptide as shown in the docked model in Figure 2a & 2b. It was observed in the predicted models that the charged and polar side chains are pointed away from the dimer while the non-polar side chains are present in between the two helices. The 2nd Val, 6th Ala and 10th Val of the 11-mer were involved in the hydrophobic interactions. We further investigated which of these positions play more important role and if there can be peptides with better binding than wild type sequence. Such knowledge can help guide designing of molecules that can bind to individual monomers and stabilize them, thus having inhibitory effect on formation of aggregates.
Docking of the 11-mer combinatorial library with SNCA
Three positions which were involved in the hydrophobic interactions were selected to be computationally mutated into Leu, Ile or Val. Two of these positions already had a Val and these were mutated only to Leu and Ile. This led to a library of 36 peptides which were used to dock on the SNCA protein molecule within the grid defining the 11-mer region. These 36 peptides along with the peptide with wild type sequence were docked on the SNCA and were evaluated based on the intermolecular energy in kcal/mol. The schematic of the procedure is shown in Figure 3. As shown by the energy histogram in Figure 4, there were several peptides which showed a better (negative) intermolecular energy than the peptide with wild type sequence (shown as blue star in Figure 4). Two peptides with lowest intermolecular energies are also shown. A common feature of these two peptides with better binding is presence of a larger hydrophobic group (Ile) in the centre of the assembly i.e. 6th position in the 11-mer peptide. This position thus seems to be important and can be potential target for inhibitory molecules designed to halt the protein aggregations in synucleinopathies involving SNCA.
Using computational approaches, we attempted to understand the initial steps of oligomerization process of SNCA. Using predicted docked models of SNCA dimers, we identified key residues involved in the interaction of the assembly. Using combinatorial approaches to mutate these key residues, we found several peptides that have better binding energies. The central hydrophobic residue is found to be most critical and potential target for inhibitory molecules. These findings may guide therapeutic strategies towards major neurodegenerative disorders involving synucleinopathies.
The authors are grateful to Dr Kota Reddy, Academic President and Dr Firdos Alam Khan, Chairperson, School of Life Sciences, Manipal University, Dubai, for their constant support and encouragement. Ms. Sana Siddiqui is acknowledged for her helpful and insightful discussions during initial stages of this work.
The author declares no conflict of interest.

©2015 Muhammad, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.