MOJ
eISSN: 2374-6920


Research Article Volume 6 Issue 4
1Department of Pathophysiology, Institute of Basic Medical Sciences Chinese Academy of Medical Sciences, China
2Department of Pathology, Capital Medical University, China
3Department of Biochemistry and Molecular Biology, Beijing Normal University, China
Correspondence: Youhe Gao, Department of Biochemistry and Molecular Biology, Beijing Normal University, Gene Engineering and Biotechnology Beijing Key Laboratory, Beijing, China
Received: November 21, 2017 | Published: November 24, 2017
Citation: Yuan Y, Zhang F, Ni Y, et al. Urinary protein changes in a rat starvation model. MOJ Proteomics Bioinform. 2017;6(4):323-331. DOI: 10.15406/mojpb.2017.06.00202
Protein has been found in urine in all relevant studies. Why are there proteins in urine? If they act as nutrients, should all proteins be reabsorbed into blood during starvation to maintain homeostasis of internal environment? Are they toxic? If they are toxic or are discarded for regulation of body's own cellular functions, they still must be released into urine even after starvation. Does kidney need proteins to maintain urine flow? If proteins are necessary to maintain urine flow, at least some of them should remain in urine after starvation. In this study, five SpragueDawley rats were starved continuously for 7days. The quantity, composition, and posttranslational modification of the urine proteome was studied before and after starvation. After 7-day starvation, urinary protein concentration had no significant changes, even though serum protein concentration decreased for about 10%. Only five and thirteen urinary proteins were significantly changed in the 4- and 7-day starvation groups respectively, compared with before starvation group. These findings indicate there were still proteins even after starvation which supports those urinary proteins may be necessary for kidney to maintain urine flow or be toxic and discarded for the regulation of cellular functions. It seems that removing proteins in urine is important for the survival of the animals.
Keywords: urinary proteins, homeostasis, toxic, kidney, starvation
Protein has been found in urine in all relevant studies. Why are there proteins in urine? If the protein in urine acts as a nutrient, should all proteins be reabsorbed into the blood during starvation to maintain the homeostasis of the internal environment? Are they toxic? Is there any protein in urine when the animal is starved? If the protein in urine is toxic or is discarded for the regulation of body's own cellular functions, it still must be released into the urine even when the animal is starved. Does the kidney need protein to maintain urine flow? If the protein is necessary to maintain urine flow, at least some of it should remain in the urine even after starvation. Starvation can lead to different disorders of the body. During the process of starvation, a series of energy metabolism changes leads to a decrease of body weight and changes in the body’s components and the metabolic gene expression profile.1,2 A short duration of starvation can cause the acceleration of protein metabolism3 and increases in nitrogen excretion, leucine flux and oxidation.4 All of the above observations illustrate that the effect of starvation on the body may possibly reflect urinary protein changes.
The metabolic response to starvation is divided into three phases according to changes in glucose, lipid and protein utilization.5 In the first phase, the glucose composition increases, and a large quantity of lipid is stored. In the second phase, the energy supply is largely from stored fat and a small quantity of protein. In the third phase, most of the protein in the body is utilized for the energy supply. In different phases, different levels of protein consumption may be reflected by different urinary protein changes. The starving phase was determined by calculating the specificDaily rate of body mass loss dM/Mdt (g/kg per d) for each animal.6,7 In this paper, Sprague-Dawley rats starved 4 and 7days were chosen as the second and third phase of starvation and urinary and serum protein changes were analyzed by liquid chromatography–tandem mass spectrometry (LC-MS/MS) to distinguish the possible effects of starvation.
Ethics statement
The animal experiments were reviewed and approved by the Institute of Basic Medical Sciences Animal Ethics Committee, Peking Union Medical College (Approved ID: ACUC-A02-2014-008). The study was performed strictly according to the guidelines developed by the Institutional Animal Care and Use Committee of Peking Union Medical College. All efforts were made tominimize suffering.
Experimental animals and starvation model
Specific pathogen-free male Sprague-Dawley rats weighing 180-200 g were purchased from the Institute of Laboratory Animal Science, Chinese Academy of Medical Science & Peking Union Medical College. Five rats were starved continuously for 7days with free access to water. Body weight and 24-h urine volume were recordedDaily. Rats were housed in metabolic cages to collect urine samples. After starvation for 4 and 7days, 500µL of blood was collected from the caudal vein of each rat.
Laboratory biochemical analysis
Urine and serum were analyzed in the clinical laboratory of Peking Union Medical College Hospital. The urinary protein and creatinine and serum protein and creatinine concentrations were measured.
Urinary protein and serum protein sample preparation
Urinary protein was extracted according the instruction.8 Serum samples were diluted for further use. Urinary protein and serum protein concentrations were measured using the Bradford method.
Proteome analysis
The urinary and serum proteins were digested with trypsin (Trypsin Gold, Mass Spec Grade, Promega, and Fitchburg, WI, USA) using filter-aided sample preparation methods.9
The digested peptides were acidified and re-dissolved in 0.1% formic acid and were then loaded onto a reversed-phase micro-capillary column using a Waters Ultra Performance Liquid Chromatography (UPLC) system. The peptides were separated on a 10-cm fused silica column and then eluted over a gradient of 5%-28% buffer B (0.1% formic acid, 99.9% ACN; flow rate, 0.3µL/min) for 100min. MS Data were acquired using an ABSCIEX Triple-TOF 5600 mass spectrometer system (Framingham, MA, US). Five animals were randomly chosen, and their urinary proteins and serum proteins from before, 4- and 7-day after starvation groups were analyzed and compared. The MS/MS Data were processed using Mascot software (version 2.4.1, Matrix Science, London, UK) and were searched against the Swiss-Prot rat Database (05/03/2013; containing 9,354 sequences). For peptide identification, the parent ion tolerance was set to 0.05Da, and the fragment ion mass tolerance was set to 0.05Da. The search allowed for two missed cleavage sites in trypsin digestion. The carbamidomethylation of cysteine was considered as a fixed modification, and both the oxidation of methionine and the deamidation of asparagine were considered as variable modifications. The Mascot results were filtered and validated using Scaffold (version 4.0.1, Proteome Software Inc., Portland, OR, USA). Peptide identifications were accepted if they were detected with ≥90.0% probability and a false discovery rate (FDR) of less than 0.1% using the Scaffold local FDR algorithm. Protein identifications were accepted if they were detected with ≥95.0% probability and contained at least 2 identified peptides.9
Spectral counting analysis for differential proteins
Spectral counting was used to compare protein abundances before and 4- and 7-day after starvation, according to the procedure described previously.10–12 The proteins with spectral counts that differed between the before starvation group and the 4- or 7-day after starvation groups above internal experimental variation were defined as changed proteins. The proteins whose spectral counts were more than 2-fold higher or lower in the 4- or 7-day starvation groups were chosen as differential proteins. In addition, the difference between significantly changed urinary and serum proteins were also analyzed and compared.
Protein posttranslational modification analysis
Posttranslational modification (PTM) of urinary proteins and serum proteins in the before and 4- and 7-day after starvation groups were analyzed and compared using Peaks software (Bioinformatics Solutions Inc., Canada).13,14 The MS/MS Data were searched against the Swiss-Prot rat Database. The parent and fragment mass error tolerance was set to 0.05Da. The search allowed for two missed cleavage sites and one non-specific cleavage site in the trypsin digestion. The carbamidomethylation of cysteine was considered to be a fixed modification. Posttranslational modifications were accepted if they were detected with an FDR of less than 0.5% by the Scaffold local FDR algorithm. Forty types of real in vivo posttranslational modifications (Table S1) of urinary proteins were checked, and six types of in vivo posttranslational modifications of fourteen target proteins were checked in both serum and urine. Furthermore, the significant change of PTMs between independent groups was selected.
Table S1 In vivo PTMs by Unimod
Statistical analysis
Comparisons of changed urinary and serum proteins in the before, 4- and 7-day starvation groups were conducted using repeated measures ANOVA. If the result was statistically significant, then the comparison of changed proteins between each of the two groups was analyzed by one-way ANOVA followed by post hoc analysis with the least significant difference (LSD) test. Group differences resulting in P-values of less than 0.05 were considered statistically significant. Statistical analysis was performed using the Statistical Package for Social Studies (SPSS) version 16.0.
Body weight and urine volume decreased for starved rats
The body weight and urine volume of rats decreased gradually during starvation. Compared to before starvation group, the body weights decreased by 14.08% and 19.37% in the 4- and 7-day after starvation groups, respectively (Figure 1a). There was a downward trend of body weight loss after starvation for 0 to 2days, it stayed in the plateau of starvation for 3 to 6days, and increased slightly after starvation for 7days (Figure 1b). In addition, the urine volume decreased 51.79% in the 4-day starvation group and 61.70% in the 7-day starvation group relative to before starvation group (Figure 1c).
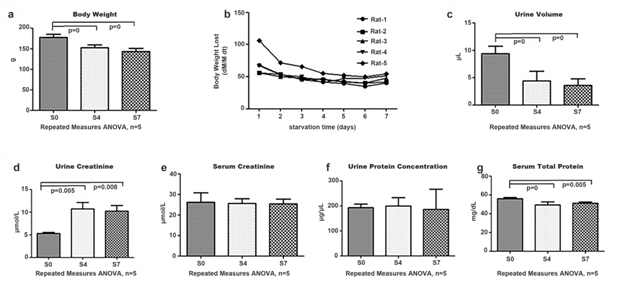
Figure 1a The body weight change after starvation. The histograms show the average body weight of 5 rats.
Figure 1b The speed of body weight lost after starvation. The different curves show the body weight lost (BWL) change of each rat. The data represent the value of BWL.
Figure 1c The urine volume change after starvation. The histograms show the average urine volume of 5 rats.
Figure 1d The urine creatinine change after starvation. The histograms show the average urine creatinine of 5 rats.
Figure 1e The serum creatinine change after starvation. The histograms show the average serum creatinine of 5 rats.
Figure 1f The urine protein concentration change after starvation. The histograms show the average urine protein concentration of 5 rats.
Figure 1g The serum protein concentration change. The histograms show the average serum protein concentration of 5 rats. The data represent mean ± SD. S0: before starvation group; S4: 4-day starvation group; S7: 7-day starvation group.
Starvation mainly led to a series of energy metabolism changes and caused decreases in body weight and differences in body components and metabolic gene expression.1,2 In previous studies, the starvation process was divided into three phases according to different main energetic materials,5 and body weight loss was used to confirm different phases. In this study, starvation for 1 to 2days was detected to be in the first phase, and the value of body weight loss decreased. During starvation for 3 to 6days, which was in the second phase, the value was almost stable. Using starvation for 7days as the third phase, the value increased slightly. In the second and third phases, the protein exhaustion profiles were different,5 and it was likely to identify changes in the urinary and serum proteome.
The serum creatinine level was not changed and the urine creatinine level increased in starved rats. The mean value of urine creatinine was 1.91-fold higher in the 4-day starvation group and 1.88-fold higher in the 7-day starvation group relative to before starvation group (Figure 1d). The serum creatinine showed no significant changes during starvation (Figure 1e).
Relative to before starvation group, the serum creatinine levels showed no significant changes, while the urine creatinine levels increased significantly in the 4- and 7-day starvation groups. The reason for this observation might be that a large quantity of muscle proteins was consumed to provide energy during these two phases, and the metabolite creatinine was released into the blood. To maintain the homeostasis of blood, creatinine was removed into urine, resulting in its increase in the urine. It agreed really well with our theory that urine is an earlier and more sensitive biomarker source than blood.15
Even the serum protein concentration level decreased in starved rats, the urinary protein concentration level had no significant change. The urinary protein concentration levels remained the same during starvation (Figure 1f) even the serum protein concentration levels decreased to 88.14% and 91.46% in the 4- and 7-day starvation groups, respectively, relative to before starvation group (Figure 1g). Serum protein concentration change was possibly due to the large quantity of protein utilization during starvation. However, that the urinary protein concentrations showed no significant changes after starvation for 4 and 7days was really a surprise.
Proteins were considered having nutritious value and theoretically should be all reabsorbed in order to maintain the stable protein concentration of the blood according to the homeostasis of internal environment theory. These findings support that urinary proteins may be necessary for kidney to maintain the urine flow or were toxic and discarded for the regulation of cellular functions for the survival of the animals.
Urinary proteome and serum proteome changes identified by LC-MS/MS
Fifteen urine samples and fifteen serum samples (taken from five rats per group) were analyzed by UPLC coupled with triple time-of-flight 5600 MS. Spectral counting was used to perform semi-quantitative analysis. In this study, the abundance of each given protein in a sample was estimated by the mean spectral count of proteins that existed in all 5 replicates.
In total, 663 urinary proteins were identified by scaffold integration. Relative to before starvation group, 5 and 13 significantly changed urinary proteins were identified in the 4- and 7-day starvation groups, respectively (Tables 1) (Table 2) (Figure 2 & 3). In total, 99 serum proteins were identified by scaffold integration. Relative to before group, 1 and 23 significantly decreased proteins were identified in the 4- and 7-day starvation groups (Tables 3 & 4). The significantly changed proteins criteria: 1) only proteins with at least two unique peptides were included; 2) the variation trend in all five animals in the 4- or 7-day starvation group was consistent, and the fold-change of the mean value of spectral counting in these two groups was >2 or <0.5 compared to that in before starvation group; and 3) the spectral counting was >5 in higher protein expression groups.
Protein Name |
Fold- |
p value |
Spectral Counts |
||||||||||||||
S0-1 |
S0-2 |
S0-3 |
S0-4 |
S0-5 |
S4-1 |
S4-2 |
S4-3 |
S4-4 |
S4-5 |
S7-1 |
S7-2 |
S7-3 |
S7-4 |
S7-5 |
|||
Cluster |
2.15↑ |
0.002 |
10 |
14 |
11 |
18 |
16 |
32 |
28 |
34 |
24 |
29 |
46 |
43 |
31 |
37 |
25 |
Annexin A4 |
2.06↑ |
0.025 |
2 |
6 |
2 |
5 |
6 |
5 |
9 |
11 |
11 |
7 |
5 |
5 |
3 |
6 |
8 |
Haptoglobin |
0.34↓ |
0.048 |
12 |
5 |
10 |
12 |
9 |
1 |
1 |
0 |
0 |
1 |
0 |
1 |
3 |
1 |
0 |
Cluster |
0.44↓ |
0.002 |
6 |
6 |
14 |
7 |
8 |
4 |
2 |
7 |
3 |
2 |
2 |
4 |
4 |
3 |
2 |
Complement component C9 |
0.45↓ |
0.023 |
6 |
5 |
10 |
7 |
8 |
2 |
6 |
2 |
1 |
5 |
3 |
2 |
1 |
5 |
2 |
Table 1 Urinary proteins displaying significant changes between the starvation 0-day and 4-day groups
Protein Name |
Fold- |
p value |
Spectral Counts |
||||||||||||||
S0-1 |
S0-2 |
S0-3 |
S0-4 |
S0-5 |
S4-1 |
S4-2 |
S4-3 |
S4-4 |
S4-5 |
S7-1 |
S7-2 |
S7-3 |
S7-4 |
S7-5 |
|||
Interleukin-1 receptor type 2 |
4.94↑ |
0 |
1 |
1 |
2 |
2 |
3 |
5 |
5 |
3 |
4 |
6 |
9 |
8 |
7 |
7 |
8 |
Apolipoprotein M |
3.36↑ |
0.001 |
4 |
3 |
5 |
6 |
3 |
8 |
6 |
9 |
5 |
4 |
14 |
14 |
13 |
7 |
14 |
Cluster |
3.00↑ |
0.002 |
10 |
14 |
11 |
18 |
16 |
32 |
28 |
34 |
24 |
29 |
46 |
43 |
31 |
37 |
25 |
Ciliary neurotrophic factor receptor subunit alpha |
2.66↑ |
0.008 |
1 |
1 |
3 |
3 |
4 |
4 |
5 |
5 |
2 |
5 |
5 |
7 |
5 |
5 |
7 |
Neutrophil gelatinase-associated lipocalin |
2.22↑ |
0.009 |
4 |
2 |
4 |
4 |
5 |
4 |
3 |
5 |
2 |
9 |
9 |
6 |
7 |
7 |
8 |
Complement C3 |
0.30↓ |
0 |
33 |
41 |
31 |
42 |
59 |
12 |
20 |
8 |
16 |
24 |
9 |
16 |
8 |
17 |
5 |
Complement component C9 |
0.41↓ |
0.023 |
6 |
5 |
10 |
7 |
8 |
2 |
6 |
2 |
1 |
5 |
3 |
2 |
1 |
5 |
2 |
Cluster of Calbindin |
0.42↓ |
0.002 |
6 |
6 |
14 |
7 |
8 |
4 |
2 |
7 |
3 |
2 |
2 |
4 |
4 |
3 |
2 |
L-lactate dehydrogenase B chain |
0.42↓ |
0.019 |
5 |
5 |
7 |
5 |
9 |
2 |
7 |
6 |
4 |
9 |
0 |
4 |
3 |
3 |
1 |
Cluster of Alpha-1B-glycoprotein |
0.44↓ |
0.005 |
14 |
11 |
17 |
10 |
12 |
7 |
10 |
6 |
7 |
7 |
3 |
5 |
4 |
3 |
10 |
C-reactive protein |
0.45↓ |
0 |
24 |
25 |
22 |
24 |
23 |
17 |
20 |
18 |
16 |
9 |
10 |
14 |
9 |
10 |
4 |
Vitamin D-binding protein |
0.47↓ |
0 |
52 |
47 |
31 |
40 |
51 |
27 |
26 |
12 |
20 |
26 |
21 |
21 |
10 |
18 |
21 |
Phosphatidylethanolamine-binding protein 1 |
0.48↓ |
0 |
9 |
12 |
17 |
10 |
11 |
11 |
14 |
15 |
18 |
17 |
4 |
4 |
5 |
6 |
6 |
Table 2: Urinary proteins displaying significant changes between the starvation 0-day and 7-day groups.
Protein Name |
Fold- |
p value |
Spectral Counts |
||||||||||||||
S0-1 |
S0-2 |
S0-3 |
S0-4 |
S0-5 |
S4-1 |
S4-2 |
S4-3 |
S4-4 |
S4-5 |
S7-1 |
S7-2 |
S7-3 |
S7-4 |
S7-5 |
|||
C-Reactive Protein |
0.5↓ |
0.0057 |
62 |
67 |
54 |
39 |
51 |
41 |
37 |
24 |
13 |
9 |
1 |
0 |
0 |
72 |
27 |
Table 3 Serum proteins displaying significant changes between the starvation 0-day and 4-day groups.
Protein Name |
Fold- |
p value |
Spectral Counts |
||||||||||||||
S0-1 |
S0-2 |
S0-3 |
S0-4 |
S0-5 |
S4-1 |
S4-2 |
S4-3 |
S4-4 |
S4-5 |
S7-1 |
S7-2 |
S7-3 |
S7-4 |
S7-5 |
|||
Alpha-1B-glycoprotein |
0.2↓ |
0.00044 |
97 |
109 |
100 |
97 |
124 |
70 |
78 |
69 |
61 |
59 |
8 |
0 |
0 |
42 |
67 |
Leukemia inhibitory factor receptor |
0.2↓ |
0.0025 |
17 |
18 |
21 |
23 |
30 |
10 |
16 |
13 |
11 |
11 |
1 |
0 |
1 |
8 |
16 |
Complement C3 |
0.3↓ |
0.0054 |
562 |
668 |
607 |
541 |
683 |
480 |
512 |
379 |
382 |
252 |
80 |
21 |
8 |
556 |
324 |
Fibronectin |
0.3↓ |
0.016 |
152 |
126 |
111 |
86 |
133 |
98 |
118 |
77 |
76 |
53 |
8 |
0 |
0 |
113 |
84 |
Complement component C9 |
0.3↓ |
0.014 |
61 |
56 |
65 |
48 |
50 |
58 |
59 |
51 |
41 |
25 |
6 |
0 |
0 |
59 |
29 |
C4b-binding protein alpha chain |
0.3↓ |
0.0092 |
30 |
28 |
25 |
22 |
28 |
22 |
24 |
19 |
14 |
12 |
3 |
0 |
0 |
27 |
12 |
Phosphatidylinositol-glycan-specific phospholipase D |
0.3↓ |
0.0067 |
14 |
10 |
8 |
11 |
9 |
14 |
15 |
6 |
12 |
6 |
2 |
2 |
0 |
9 |
5 |
Insulin-like growth factor-binding protein complex acid labile subunit |
0.3↓ |
0.025 |
19 |
22 |
9 |
12 |
8 |
12 |
9 |
7 |
5 |
5 |
0 |
0 |
0 |
13 |
5 |
Complement C1s subcomponent |
0.3↓ |
0.011 |
9 |
9 |
11 |
6 |
11 |
12 |
12 |
10 |
4 |
3 |
2 |
1 |
0 |
9 |
4 |
Extracellular matrix protein 1 |
0.3↓ |
0.023 |
9 |
7 |
8 |
8 |
8 |
11 |
8 |
13 |
8 |
3 |
0 |
0 |
0 |
9 |
5 |
Hemopexin |
0.4↓ |
0.039 |
262 |
260 |
217 |
217 |
311 |
260 |
262 |
213 |
199 |
181 |
76 |
11 |
1 |
259 |
223 |
Complement C4 |
0.4↓ |
0.035 |
149 |
176 |
139 |
139 |
164 |
162 |
176 |
110 |
111 |
91 |
48 |
15 |
2 |
180 |
99 |
Apolipoprotein A-I |
0.4↓ |
0.022 |
82 |
96 |
59 |
62 |
99 |
83 |
100 |
65 |
59 |
46 |
15 |
2 |
0 |
79 |
52 |
Afamin |
0.4↓ |
0.029 |
100 |
97 |
72 |
74 |
97 |
82 |
88 |
64 |
65 |
37 |
9 |
2 |
0 |
82 |
81 |
Vitamin D-binding protein |
0.4↓ |
0.032 |
99 |
87 |
80 |
77 |
95 |
103 |
86 |
72 |
46 |
36 |
9 |
3 |
1 |
103 |
57 |
C-reactive protein |
0.4↓ |
0.048 |
62 |
67 |
54 |
39 |
51 |
41 |
37 |
24 |
13 |
9 |
1 |
0 |
0 |
72 |
27 |
Protein AMBP |
0.4↓ |
0.032 |
30 |
30 |
28 |
31 |
33 |
32 |
27 |
26 |
28 |
10 |
5 |
0 |
0 |
32 |
26 |
Serum paraoxonase/arylesterase 1 |
0.4↓ |
0.032 |
27 |
22 |
25 |
23 |
30 |
29 |
21 |
18 |
17 |
15 |
5 |
1 |
0 |
28 |
19 |
Clusterin |
0.4↓ |
0.018 |
27 |
22 |
23 |
17 |
29 |
29 |
26 |
19 |
13 |
9 |
2 |
1 |
0 |
23 |
16 |
Apolipoprotein M |
0.4↓ |
0.016 |
11 |
11 |
11 |
10 |
10 |
14 |
11 |
11 |
10 |
7 |
3 |
2 |
0 |
11 |
7 |
Transthyretin |
0.4↓ |
0.034 |
15 |
13 |
12 |
13 |
13 |
12 |
13 |
9 |
10 |
10 |
5 |
0 |
0 |
15 |
9 |
Coagulation factor X |
0.4↓ |
0.02 |
12 |
10 |
9 |
9 |
8 |
8 |
10 |
7 |
6 |
5 |
4 |
0 |
1 |
11 |
2 |
Attractin |
0.5↓ |
0.04 |
10 |
11 |
12 |
11 |
13 |
11 |
9 |
8 |
6 |
3 |
3 |
0 |
2 |
14 |
7 |
Table 4 Serum proteins displaying significant changes between the starvation 0-day and 7-day groups
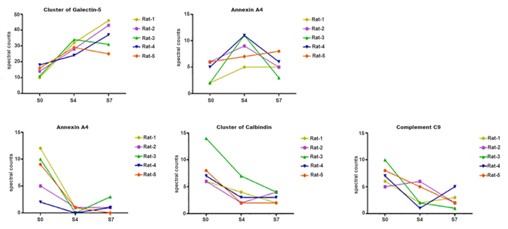
Figure 2 Changed urinary proteins after starvation 4 days. The curves show the increasing trend of 2 changed proteins of 5 rats in starvation 0, 4 and 7 days. S0: starvation 0-day group; S4: starvation 4-day group; S7: starvation 7-day group.

Figure 3 Changed urinary proteins after starvation 7 days. The curves show the increasing trend of 5 changed proteins of 5 rats in starvation 0, 4 and 7 days. S0: starvation 0-day group; S4: starvation 4-day group; S7: starvation 7-day group.
During the starvation process, the levels of some proteins increased in urine, such as apolipoprotein M. Increased concentrations of apolipoprotein M has been observed with the utilization of fat.16,17 Apolipoprotein M is a novel type of apolipoprotein that bonds to high-density lipoprotein. The over-expression of this protein could lead to lipid consumption and eventually induce the reduction of plaques in the aortic root.17 Therefore, when starvation caused different degrees of fat consumption, the levels of these Apolipoprotein M in the urine of the 7-day starvation group increased.
During the starvation process, the levels of some proteins decreased in urine, which might be reabsorbed by the body or leached out by glomerular filtration, such as complement C3, complement component C9 and C-reactive protein. These proteins play important roles in many vital activities in the body, such as immune reactions and glycolysis.18,19 In the study of very young obese children, vitamin D-binding protein has been observed to decrease the reduction of body weight.20 In a word, the proteins whose levels decreased during the starvation process were nutrient proteins and might be reused more to maintain the relative stability of the body.
Comparing 8 significantly decreased urinary proteins to serum proteins in the 7-day group starvation, complement C3, complement component C9, C-reactive protein and vitamin D-binding protein experienced the same downtrends in serum. The decreased levels of these four proteins in urine may be caused by their corresponding decreases in serum, producing a “funnel effect”: in a state of starvation, protein sources are reduced; therefore, protein excretion in the urine is also reduced. The other four proteins were not identified in the serum proteome without the removal of highly abundant proteins, which indicated the low abundance of these proteins in serum. These proteins may be used to maintain homeostasis in the serum. Among the five significantly increased urinary proteins, apolipoprotein M had a different decreasing tendency in serum, while interleukin-1 receptor type 2, cluster of galectin-5, CNFR-α and neutrophil gelatinase-associated lipocalin were also not identified in the serum proteome without the removal of highly abundant proteins. The Data reported here suggested that in a state of starvation, even though the sources of proteins were reduced, the excretion of these five proteins in the urine was still increased, which may indicate the useless or harmful nature of these proteins. In addition, the levels of afamin, attractin, clusterin, complement C4, fibronectin, hemopexin, leukemia inhibitory factor receptor, protein AMBP and transthyretin were decreased in serum; however, no significant changes were found in urine. It seems that in spite of the decreases in certain serum proteins in starvation, protein excretion was not changed. However, the results only show the comparison of urinary proteins’ and serum proteins’ changing trends in starvation, and further research is needed to understand the mechanism of the proteins’ filtration and whether protein in urine due to regulation of body's own cellular functions.
Kidney injury markers were not present in urine of starved rats
Proteins were still in urine at almost the same concentration, even though the serum protein concentrations decreased and the serum’s homeostatic internal environment was disrupted. Is this against the homeostasis of internal environment? Moreover, some proteins were unchanged or even increased in urine despite their decrease in serum in the 7-day starvation group. Were proteins present in urine due to kidney injury? Many kidney injury urinary biomarkers have been reported by proteomic analyses in recent years,21–26 among which some have also been investigated in animal models, such as nestin22 and vimentin;24 however, most of these markers showed no significant changes in starvation status. These results indicated that proteins were still excreted in urine in starvation rather than the result of kidney injury.
Posttranslational modification changes in urinary and serum proteins by peaks
Posttranslational modification refers to the attachment of biochemical functional groups to one or more amino acids of a polypeptide chain before it becomes a mature protein product. Proteins undergoing posttranslational modification play important roles in cell signaling and the modulation of enzyme activity.27 In this study, changes in vivo posttranslational modifications of urinary proteins were observed. These observations may be important for the understanding of different changes caused by starvation.
The changed PTMs on a fixed site in each group were analyzed, and the significantly changed PTMs were screened. The screened criteria included the following: 1) the detected number of a specific PTM in each animal in the starvation group was >5; and 2) the variation between each group was statistically significant (p<0.05). Compared to before starvation group, urinary protein formulation and phosphorylation were significantly changed in the 7-day starvation group (Table 5). To detect the different PTMs between serum and urinary target proteins when starving, six in vivo posttranslational modifications of fourteen proteins were compared between serum and urinary proteins in 7-day starvation group (Table 6) (Figure 4). However, no regular changes were observed.
Name |
Formylation |
Phosphorylation |
∆Mass |
27.99 |
79.97 |
location |
K,N-term |
STY |
S0-1 |
32 |
29 |
S0-2 |
19 |
22 |
S0-3 |
20 |
19 |
S0-4 |
30 |
40 |
S0-5 |
27 |
26 |
S4-1 |
26 |
28 |
S4-2 |
22 |
32 |
S4-3 |
25 |
27 |
S4-4 |
24 |
32 |
S4-5 |
34 |
33 |
S7-1 |
29 |
23 |
S7-2 |
8 |
12 |
S7-3 |
14 |
16 |
S7-4 |
14 |
12 |
S7-5 |
16 |
17 |
Change |
↓ |
↓ |
p value |
0.034 |
0.008 |
Table 5 PTMs displaying significant changes between the Starvation 0-day and 7-day groups
Protein Name |
In Urine |
In Serum |
Apolipoprotein M |
↑ |
↓ |
Afamin |
— |
↓ |
Attractin |
— |
↓ |
Clusterin |
— |
↓ |
Complement C4 |
— |
↓ |
Fibronectin |
— |
↓ |
Hemopexin |
— |
↓ |
Leukemia inhibitory factor receptor |
— |
↓ |
Protein AMBP |
— |
↓ |
Transthyretin |
— |
↓ |
Complement C3 |
↓ |
↓ |
Complement component C9 |
↓ |
↓ |
C-reactive protein |
↓ |
↓ |
Vitamin D-binding protein |
↓ |
↓ |
Table 6 Change tendency of fourteen target proteins in urine and serum
↑: increased significantly; ↓: decreased significantly; —: no significant difference
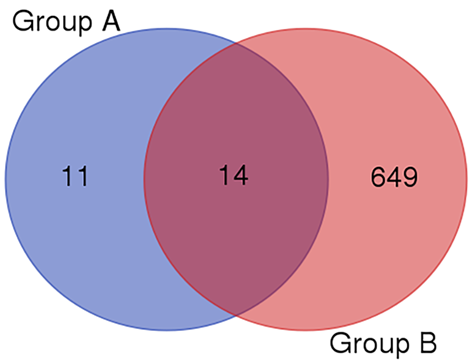
Figure 4 Target proteins. Group A: Significantly decreased serum proteins in starvation 7-day group; Group B: Total urinary proteins.
To identify the reason why proteins are present in urine even in starvation status, the PTMs of urinary serum proteins was also analyzed. However, no significant regularity was found. Results indicated that the existence of protein in urine were not caused by posttranslational modifications. However, the current results are not sufficient to reach this conclusion. Due to the different changes of composition and quantity of urinary and serum proteins in starvation, the amount of PTM sites may be affected as well, which may have an influence on the comparison of urinary and serum protein posttranslational modification. Further studies are still needed to understand the reason and meaning of the different posttranslational modifications.
Proteins are still in urine, even though the serum protein concentrations decreased in starvation status, which against the theory of homeostasis of internal environment. It implies that for the survival of the animals, urinary proteins may be necessary for kidney to maintain the urine flow or were toxic and discarded for the regulation of cellular functions. Some of the changes proteins identified in this study may be potential biomarkers for evaluating the degree of starvation. Further studies are still necessary to explain the exact reason why proteins exist in urine even after starvation and what are the markers of the toxic proteins.
T his work was supported by the National Key Research and Development Program of China (2016YFC1306300); the National Basic Research Program of China (2013CB530850); Beijing Natural Science Foundation (7173264, 7172076) and funds from Beijing Normal University (11100704, 10300-310421102).
The author declares no conflict of interest.

©2017 Yuan, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.