MOJ
eISSN: 2374-6920


Research Article Volume 5 Issue 3
Department of Computer and Electrical Engineering and Computer Science, Florida Atlantic University, USA
Correspondence: Mirjana Pavlovic, Research Professor, Department of Computer and Electrical Engineering and Computer Science., Florida Atlantic University, 777777 Glades Rd, Boca Raton, Fl 33431, USA, Tel 5612972348
Received: January 27, 2017 | Published: April 6, 2017
Citation: Kats A, Pavlovic M, Chen R, et al. Comparison and analysis of different methods for purification of autoimmune antibody reactive with single stranded DNA: a pilot study. MOJ Proteomics Bioinform. 2017;5(3):94-107. DOI: 10.15406/mojpb.2017.05.00161
Purpose: We have compared a two-step affinity purification method, previously developed in house, which utilizes biotinylated oligo-deoxythymidine (dT) bound streptavidin (SA) M-280 magnetic beads and protein G Dynabeads®, with Melon™ Gel, another two step commercial method, for the isolation and purification of human anti-DNA autoantibodies reactive with single stranded DNA (ssDNA), in order to determine which method is more applicable for analysis of antibody subclasses and, potentially subclass functional activities.
Results: Although Melon Gel allowed for faster anti-ssDNA autoantibody purification and higher recovery rate, its final product was of lower purity than that of the magnetic bead method, as confirmed with nanogram silver staining method following PhastGelTM non-reducing SDS-PAGE. The polyclonal nature of anti-ssDNA autoantibodies produced by patients with Systemic Lupus Erythematosus (SLE) has been determined and compared with normal human, and B-Chronic Lymphocytic Leucosis- (B-CLL) anti-ssDNA autoantibodies. Characterization of isolated antibodies by isoelectric focusing, western blot analysis and ELISA confirmed them to be human IgGs and detected the presence of all four IgG antibody subclasses with different participation. Additionally, Lab-on chip (Agilent 2100) method, revealed the presence of different MW patterns within lupus IgG subclasses, not detectable by SDS PAGE, which were not consistent with patterns seen in sera of control or in individuals with B-CLL.
Conclusions: The results obtained suggest incomparably better purity of antibodies isolated via the magnetic bead method vs. Melon-gel. SDS-PAGE and Lab-on-chip analyses of antibodies revealed the presence of different IgG patterns within human lupus anti-ssDNA autoantibody subclasses than those observed in healthy individuals and B-CLL patients. These patterns may be associated with SLE disease pathogenesis and highlight the importance of further molecular, structural, and functional studies of lupus anti-ssDNA antibodies.
Keywords: SLE, CLL, magnetic beads, melon gel, anti-DNA autoantibody, IgG analysis, DNA hydrolysis
The hallmark of systemic lupus erythematosus (SLE) is the production of an array of IgG and IgM auto-antibodies directed against one or more nuclear components, most commonly double stranded (ds) DNA and/or single stranded (ss) DNA. Both anti-ssDNA and anti-dsDNA antibodies are considered to be involved in disease development based on the fact that both have been eluted from the kidneys of experimental murine models and SLE patients.1 The level of anti-DNA antibodies, both ss and ds, varies in the plasma of different SLE patients, with increased levels coinciding with flares.2–4 Consequently, the level of anti-DNA antibodies in patients’ sera is used to monitor disease activity and progression.3–5 It is still not quite clear which fractions between these two categories of anti-DNA auto-antibodies are pathogenic and why.
Anti-double stranded DNA antibody, considered to be a hallmark of lupus disease, is found in 70-90% of patients with SLE (particularly in those with nephritis), and measurements of its levels in patients’ plasma is used to follow the course of disease. However, since anti-ssDNA antibody has been shown in a limited number of experimental and clinical studies to be both hydrolytic and nephritogenic, it is suggested that it may serve as a strong flare predictor.2–4 The important role of anti-ssDNA antibody is supported by studies in mouse models of nephritogenic lupus in which only anti-ssDNA antibodies were found6,7 as well as by the findings of Swanson et al.,1 & Spatz et al.,8 that some anti-dsDNA human antibodies are not pathogenic at all. The ultimate goal of this study was to determine the optimal conditions for analysis of the types and activity of highly purified human anti-ssDNA (IgG antibody) in order to gain deeper insight into functional characteristics such as binding, DNA hydrolysis, cytotoxicity and the role of these antibodies in SLE pathogenesis. This information may eventually assist the development of a method for producing highly purified anti-DNA antibodies for use in engineering dendritic cell based vaccine as well.9–15
The fundamental objection to previous attempts at purifying anti-DNA antibodies directly from human plasma16 was that the process yielded numerous bands on electrophoretic separation inconsistent with the molecular weights of IgGs. Furthermore, there was no attempt to separate or distinguish between the antibodies against ssDNA and those against dsDNA, so that the relative importance of the pathogenic role the two types play in SLE or their predictive role in the flare could be determined.2,3 Therefore, the development of a specific method for isolation and purification of each of the two categories of anti-DNA antibodies with the highest level of purity was desirable. The method would allow studies of the unique functional activities of the two types of antibodies as well as their specific role in the etio-pathogenesis of SLE and other autoimmune diseases. Because antibodies can form complexes with other proteins, it is important to prove that the catalytic and/or cytotoxic activities ascribed to anti-DNA antibodies are not due to a contamination with DNAse or some other hydrolytic protein but rather are intrinsic, constitutive properties of the antibody alone.16–18 Table 1 summarizes the similarities and differences between anti-ds and anti-ssDNA antibodies.
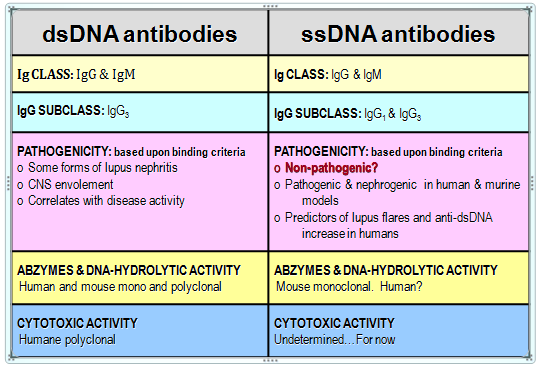
Table 1 Similarities and differences between anti-ss and anti-ds-DNA autoantibodies
Detected in diseases like SLE, RA, leukemia, lymphoma, hepatitis and nephrites. Predictors of renal failure: Diabetes mellitus type 1 and 2 Unhealthy pregnancy and preeclampsia
To our knowledge, our lab was the first to develop a specific and simple method for isolating and purifying human anti-DNA (IgG) antibodies from the serum of SLE patients19,20 based on the specific binding of anti-DNA antibodies to thymine polymers (anti-DNA antibody base specificity is dT>>dG>>dC≥dA).1,21–23 X-ray crystallography data confirmed that arginine groups are responsible for sequence recognition and anti-ssDNA antibody binding to DNA at thymine repetitive sequences. Almost an entire decade of X-ray crystallographic studies performed by Herron et al.,24 combined with the most recent data generated by Tanner in 2001,2004 and 200725,26 confirm that tyrosine and tryptophan residues create a hydrophobic pocket within the side chain of the antibody.21,22 The thymidine polymer slides between the tyrosine and tryptophan residues and while 5 consecutive thymines are necessary for recognition by arginine, only three are necessary for binding. These findings provided the basis for isolation of anti-ssDNA antibodies specific for a single strand of thymine nucleotides; using biotinylated oligo-(dT) 20mer bound to streptavidin (SA) coated magnetic Dynabeads® from InvitrogenTM (Carlsbad, CA).
The purpose of this study was to compare and contrast our beads-based method for anti-DNA antibody affinity purification19,20 with oligo-(dT) M-280 magnetic Dynabeads® and protein G Dynabeads®” to Melon™ Gel IgG Spin Purification for anti-DNA (IgG) antibody, by optimizing the conditions for anti-ssDNA (IgG) isolation and purification. This is necessary in order to achieve the maximum yield and recovery rate of antibody with the highest level of purity, and consequently, maintain the maximal functional activity during long term storage. Once purified, antibodies were analyzed via non-reducing SDS-PAGE, isoelectric focusing, Lab-on chip Agilent 2100, and ELISA in order to identify patterns which could serve as guidelines for determining their pathogenic potential in lupus. Screening of their hydrolytic activity was performed using UV light spectrophotometer and Lab-on-chip Analysis with ss DNA as the substrate.
Lupus patients, B-CLL patients and normal donors
Whole blood was intravenously drawn with written consent and under the guidance of protocols approved by the Florida Atlantic University’s Institutional review board. The plasma of lupus, B-CLL patients and normal donors was obtained by centrifuging the whole blood at 300xg for 10minutes to sediment the peripheral blood mononuclear fraction from the plasma. Supernatant fluid containing plasma was harvested and centrifuged at 600xg for 15minutes, in order to subsequently sediment and remove blood platelets. The serum was stored frozen at –20°C for future antibody purifications.
Isolation and purification methods
Anti-DNA antibody affinity purification with oligo-(dT) M-280 magnetic dynabeads® and protein G dynabeads®: A two-step design for anti-DNA antibody isolation and purification
Affinity purification of anti-DNA antibodies using Melon™ Gel IgG Purification System
The ImmunoPure® Melon™ Gel IgG Spin Purification Kit was obtained from Thermo Fisher Scientific/ Pierce Chemical (Rockford, IL). 100µl of settled Melon™ gel has the capacity to purify up to 100µl of serum. Melon™ Gel IgG Purification Support and Purification Buffer were equilibrated to room temperature and 500µl of gel slurry was dispensed into a Handee™mini-Spin Column placed in a microcentrifuge tube. The uncapped column assembly was centrifuged at 6,000xg, flow-through was discarded and the column was washed twice with 300µl of Purification Buffer. The 10-100µl of serum sample diluted 1:10 was then added to the column, incubated for 5minutes, and centrifuged at 6,000xg for 1minute; purified antibody was collected at the bottom of the collecting tube. Purified antibody was stored in Melon gel Purification buffer for further analysis or additional purification by DNA cellulose. The principles of both methods compared with past methods are visualized in Figures 1a-1c (Beads, Melon-Gel and Old-dsDNA isolation method).

Figure 1a Anti-DNA antibody affinity purification with oligo-(dT) M-280 magnetic Dynabeads and protein G Dynabeads: A two-step design for anti-DNA antibody isolation and purification.

Figure 1b Spin-column Procedure for IgG Antibody Purification.

Figure 1c Affinity purification of anti-DNA antibodies using Single stranded/Double stranded DNA-cellulose
Protein concentration determination
Protein assay with pierce micro BCATM protein assay reagent kit: A standard human IgG (10 mg/ml) was diluted to obtain 40µg/ml, 20µg/ml, 10µg/ml and 5µg /ml as standard concentrations. A serial dilution of purified human anti-DNA antibody of unknown concentration was made. 150µl each of standard human IgG solution and unknown samples were added to a microplate in triplicates. The same amount of the BCA working reagent (150µl) (Thermo Fisher Scientific/ Pierce Chemical Co., Rockford, IL) was added to each well and the solution was then mixed thoroughly on a plate shaker for 30 seconds following incubation at 37°C for 2hours. After incubation, absorbance was measured at 570nm on a plate reader. All OD readings for standard human IgG and unknown samples were subtracted from the blank reading.
Quantitative ELISA for anti-DNA antibodies in patient’s sera and determination of specific binding of anti-ssDNA antibody to poli-(dT): Anti-DNA antibody levels in normal individuals, SLE and CLL patients’ sera were measured using Oligo-(dT) 20mer (Eurofins MWG Operon, Huntsville, AL) coated Streptavidin microplates (Roche Diagnostics, Indianapolis, IN) for ELISA assay developed in our lab.19,20 ELISA quantification was conducted as previously described for quantification of antibodies in human plasma by using a four-parameter-log model from the plate reader software.23 The ELISA was also used to check the binding activity of purified anti-DNA antibody.
Lab-on-chip Agilent 2100 Bioanalyzer for molecular weight and antibody purity and concentration determination
Principles of protein analysis on a chip: The Agilent electrophoretic assays are based on traditional gel electrophoresis principles that have been transferred to a chip format. The chip format dramatically reduces separation time and sample consumption. The system provides automated sizing and quantitation information in a digital format (Agilent Technologies, Santa Clara, CA).
Charged biomolecules like DNA or RNA and proteins are electrophoretically driven by a voltage gradient-similar to slab gel electrophoresis. Because of a constant mass-to-charge ratio and the presence of a sieving polymer matrix, the molecules are separated by size with smaller fragments migrating faster than larger ones. Fluorescent dye molecules intercalate into DNA and RNA strands or Protein-SDS micells. These complexes are detected by laser-induced fluorescence. Data is translated into gel-like images (bands) and electropherograms (peaks). With the help of a ladder that contains fragments of known sizes and concentrations, a standard curve of migration time versus fragment size is plotted. Size is calculated from migration times measured for each fragment in the sample. Two protein marker fragments were run with each of the samples bracketing the overall sizing range. The “lower” and “upper” markers are internal standards used to align the ladder data with data from the sample wells. This is necessary to compensate for drift effects that may occur during the course of a chip run. For protein assays, quantitation is done with the help of the upper marker. The area under the upper marker peak is compared with the sample peak areas. Because the concentration of the upper marker is known, the concentration for each sample can be calculated. Besides this relative quantitation, an absolute quantitation is available for protein assays, using external standard proteins.
SDS-PAGE electrophoresis for antibody purity and electrophoretic pattern determination
SDS-PAGE (PolyAcrylamide Gel Electrophoresis) was performed using the Pharmacia PhastSystemTM and PhastGelTM Gradient 4-15 separation gels, PhastGelTM buffer strips and full-range rainbow molecular weight markers (GE Healthcare, Piscataway, NJ). The protein separation procedure was completed by following the PhastSystemTM Owner’s Manual (Separation Technique File No.130). The detection range of the gradient 4-15 separation gel is 30 to >300KD. Any protein with a molecular weight below 30KD may appear on the stained gel, but the migration distance may not be in linear proportion to the log of the molecular weight. According to the protocol a molecular weight as low as 10KD can be detected on the gel and can be observed using the rainbow markers, ranging from 10 to 250KD molecular weight. The gels were silver stained to detect proteins at the nanogram level.19,20
PhastGelTM Semi-dry Isoelectric focusing (IEF) for evaluation of antibody clonality
To perform an IEF analysis, the samples of purified antibodies were applied to a gel (polyacrylamide) with electric current applied to create migration using the Pharmacia PhastSystemTM and PhastGelTM IEF Gradient 3-9 separation gels, PhastGelTM buffer strips and full-range rainbow molecular weight markers (GE Healthcare, Piscataway, NJ). The proteins in the mixture migrate according to charge density until they reach that part of the gel with a pH corresponding to their pI (PhastSystemTM Separation Technique File No. 100). At this point, their net charge is zero and migration stops. Most proteins have a pI value of between 5 and 9 depending on the number of positive and negative charges exposed to solvent. This method allows us to distinguish between mono-and polyclonal nature of anti-DNA antibodies in lupus. The gels were sliver stained, as already described19,20 and visualized at a nanogram level in the range 10-100ng.
Western blot for confirmation of antibody subclasses
Western blot was used to determine the identity of an electrophoretically separated protein and to measure relative amounts of the protein present in different samples, based upon densitometry. Proteins (antibodies) were separated by gel electrophoresis, using PhastGelTM SDS-PAGE (4-15) gradient system (GE Healthcare, Piscataway, NJ) and then transferred to a PVDF membrane (PhastSystemTM Development Technique File No.220). The proteins (antibodies) retain the same pattern of separation seen on the gel. The blot was incubated with a generic (milk) protein to bind to any remaining open binding sites on the membrane. Mouse anti-human antibody solution was incubated with the gel for 1hour. After washing the membrane in buffer to remove unbound mouse anti-human, a secondary goat anti-mouse antibody was added. The secondary antibody is conjugated to the enzyme (horseradish peroxidase). The location of the membrane bound antibody conjugate is revealed by color development upon incubation with a colorless substrate.
Human IgG subclass profile ELISA kit
The concentrations of IgG1, IgG2, IgG3, and IgG4 subclasses in purified anti-DNA antibody preparations were determined using a commercial human IgG subclass profile ELISA kit. The Zymed® kit (InvitrogenTM, Carlsbad, CA) is a sandwich type ELISA using a horseradish peroxidase detection system. A coated microtiter plate captures monoclonal reagents which are specific to the various human IgG subclasses. The monoclonal antibodies in turn capture the human IgG subclasses, for which they are specific, out of the serum sample. The captured human IgG is then labeled with a horseradish-peroxidase anti-human IgG reagent. The detection signal is then generated in proportion to the amount of human subclass antibody.
UV spectrophotometry and Lab-on chip Agilent 11 000 determination of anti-ssDNA isolated antibody hydrolytic activity
Only one purified anti-ssDNA antibody was mixed with solution containing Mg++ and Ca++ at the optimal abzyme: substrate ratio and OD was determined after 12hrs of incubation at 280nm. The same ratio was used for Agilent nucleotide formation detection after 12hours of incubation for antibody and short (30)min incubation for DNAse 1. The banding patterns obtained on a gel were used as a criterion for estimation and evaluation of ss DNA hydrolysis.
The anti-ssDNA cellulose method yielded no results and so its use in the study was discontinued. Either binding or elution was not standardized to appropriate controls within this system. The degree of purity and molecular weight of isolated and purified anti-DNA antibodies by methods a) and b) was initially determined by SDS–PAGE and silver staining (Figures 2a & 2b). Our results have shown antibody of a very high purity obtained by magnetic bead method detected by silver staining at a nanogram level of sensitivity (after SDS-PAGE and Agilent 2100). Melon gel purifies all IgG’s, as well, but at much lower purity level, as determined by both SDS-PAGE silver staining and Lab-on-chip Agilent 2100 method (Figure 2b & 3). Numerous bands can be seen in melon-gel purified and processed samples which are not in agreement with the MW of IgGs.

Figure 2 Electrophoretic analysis of the purity of anti-DNA antibody following two different purification methods.
Purified anti-DNA antibodies from normal individuals and SLE patients are detected using Silver-Stain SDS-PAGE gel in non-reducing conditions. A. Anti-DNA antibodies purified by two-step affinity method employing magnetic beads. B. IgG’s purified by Melon™ Gel IgG Purification System.
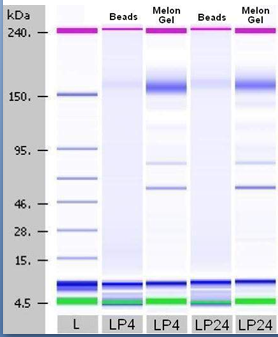
Figure 3 Agilent 2100 banding patterns of anti-ssDNA antibodies purified by SA-oligo-(dT)/ Protein G Beads versus Melon™ Gel IgG Purification.
The ladder, used to calibrate the system also provides accurate and reproducible sizing. It is clear that two-step magnetic bead affinity purification method is superior in comparison to Melon-Gel purification..
The recovery rate, although initially low, being of 20-40% can be proportionally increased by adding more beads and using our modified second step with elution buffer pH adjusted to 5, instead of 7, as previously done (19,20 - data not shown).
An initial comparison (SDS-PAGE) of anti-DNA antibodies purified from normal donors and B-cell chronic lymphocytic leukemia (BCLL) patients using two-step affinity purification method employing magnetic beads, revealed no apparent differences in the molecular weight and the number of bands. However, purified anti-DNA antibodies from the sera of patients with systemic lupus erythematosus (SLE) expressed electrophoresis patterns that differed from normal controls and varied between individual lupus patients from one-banded to four-banded.20–29 Numerous bands indicate that some proteins other then IgGs co-elute with the antibody’s fractions, during Melon Gel purification. The same conclusion can be applied to results of the Lab-on the Chip Agilent method (Figure 2b) (Table 4) where it is evident that LP4 and LP24 for example, show clear bands after magnetic beads isolation of about 150KDa, while Melon Gel purified antibody bands are “followed” by several bands of lower molecular weight indicating impurities in the final product of purification. At the same time, the number of peaks is six and eight times higher in those two representative samples of SLE patients (LP4 and LP24), although the MW of IgGs are similar, with similar percentage in the first sample and higher in the sample purified via magnetic beads (100% versus 57, 6) (Table 2). This really requires a larger sample size in order to be critically considered, but indicates that better purification of IgGs is achieved with the beads. Table 3 combines elecrophoretic and Agilent data in terms of number of bands, MW and concentration of the samples. Additionally, Agilent 2100 Lab-on-chip method confirmed the range of higher MW (164.3-234.7) in anti-DNA antibodies purified form patients with lupus, as compared to normal human IgG(157.9-178.8) and B-CLL (227.2) (Table 3). However, this method revealed a “hidden pattern” previously not detected by SDS-PAGE electrophoresis due to concentration limitations required for visualization by silver staining.29 Table 3 shows that lupus patients have a broad range of banding patterns (1-4) while normal donors and B-CLL patients have a similar pattern, i.e. two bands. Might this mean that clinically silent anti-DNA positive individuals and B-CLL patients have similar B-cell producers? Melon gel spin purification presented similar banding patterns for all normal controls, and lupus patients on both SDS-PAGE and Lab-on-chip Agilent 2100 which is to be expected (Figure 2b & 2c) (Table 3).

Figure 4 Western Blot analysis of purified anti-DNA antibodies of normal individual and SLE patients.
Western blot of a similar gel shown in figure 1A, but developed using goat anti-human IgG. Lane 1: Human IgG standard. Lanes 2,3: Lupus anti-DNA antibodies purified by Melon™ Gel IgG Purification System. Lanes 4 to 6: natural and lupus anti-DNA antibodies purified by two-step affinity method employing magnetic beads.

Figure 6 Properties of the four Human igG Subclasses literary survey.
Survey on published reports relating to general properties of the four human IgG subclasses (Calbiochem®)
Western blot analysis confirmed that the antibodies purified by both magnetic beads and melon gel spin purification, do belong to the IgG isotype (Figure 4).The striking difference is again in the purity; the comparison of the two methods reveals and confirms the superior purification efficiency of two-step magnetic beads method.
The spectrum of concentrations of antibodies after purification is shown in (Figure 5) (Table 4). With the exception of the last sample in (Table 4) (Figure 5) Melon Gel gave much higher readings for each sample concentration indicating the presence of impurities and less efficient methodological “work”. The high protein content in the last sample purified via magnetic beads (475ug/ml versus 96ug/ml), a clinically asymptomatic individual, could be due to non-efficient washing, or some external contamination with proteins.
The ELISA test for determination of antibody concentration showed a broad range of CV% (1.785-23.01); therefore the results of ODs are inconclusive (Table 5) (Figure 6).
Human IgG subclass profile ELISA detected the presence of all four IgG subclasses in both beads and Melon –gel purified samples (Figure 7 & 8). The values expressed in ug/ml were considerably higher in Melon –gel purified samples (over 1600 for IgG1 versus 46 in the bead samples), with similar participation of Igg1, IgG2 and IgG3 and difference in IgG4 being much higher in the Melon-Gel purified group. Since it is the same sample, and two different methods, we believe that higher value in Melon-gel isolated IgG4 is due to technical problem with pipeting that has created a false increase. These data are compared to the referent values for antibody subclass (Figure 8) (Table 6). As shown in Figure 7, the percentage participation in bead-isolated antibody subclasses is similar. The same was true of the PI range that will be shown later on.
Isoelectric focusing revealed the polyclonal nature of the human lupus purified antibody (a smeared patterns, pI~9) compared to mouse monoclonal antibody (three clearly distinctive bands pI~7) (Figure 10a & 10b). The data suggests that the purified antibodies from the lupus patients are polyclonal and that the B-CLL and normal donor antibodies were monoclonal, thus accounting for the different banding patterns. This also indicates that B-cells in B-CLL patients and those in normal individuals are closer in evolution than lupus B-cells.

Figure 9 Survey on published reports relating to general properties of the four human IgG subclasses (Calbiochem®).
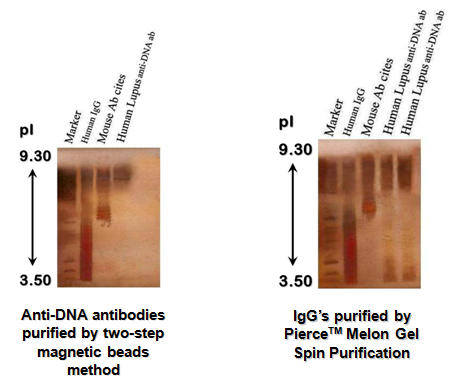
Figure 10 Purified anti-DNA antibodies from normal individuals and SLE patients are detected on Silver-Stain IEF 3-9 gel.
Figure 11 displays the results of UV spectrophotometric analysis for antibody ssDNA hydrolytic activity performed on anti-ssDNA antibody isolated from normal individuals and lupus patients via the magnetic-bead method. There is no significant change in OD in normal samples, however there is a drastic increase after 3h incubation in the lupus patient antibody hydrolytic activity.
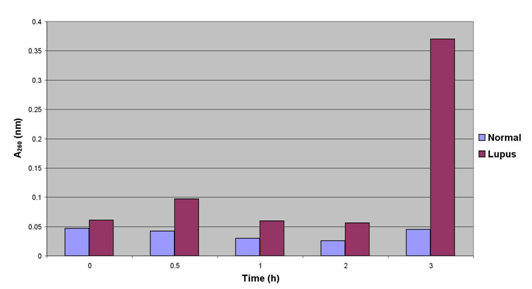
Figure 11 UV spectrophotometer performed at +4 °C, (a temperature at which DNAse 1 is inactive) also demonstrates hydrolytic activity of lupus anti-ssDNA antibody (increase in absorbance after 3 hours).
Discontinuous measurement of hydrolysis of oligo 18-mer by Lupus patient (LP) versus Normal donor (ND) by UV spectrophotometry at 260nm, 1ug/ml.
We used UV Spectrophotometry, to initially visualize hydrolytic activity of lupus anti-ssDNA antibody compared to that of a normal donor. You can see an increase on absorbance after 3 hours of lupus autoantibody hydrolysis. (Figure shows hydrolyisis of oligo 18- mer by lupus vs. normal donor also demonstrates hydrolytic activity of lupus anti-ssDNA antibody.)
Agilent -Lab-on-chip- data (Figure12a & 12b) confirmed the initial hydrolytic data obtained by UV spectrophotometry. We can see from Figure 12a that the DNAse1 reaction occurred very rapidly (about 30min necessary to obtain small (20-50bp) nucleotides starting from about 400bp herring sperm -ssDNA as the substrate). However, (Figure 12b), about 12h of incubation at 37°C degrees was necessary to hydrolyze the same substrates to small nucleotides by anti-ssDNA antibody.
We did not compare Melon-gel isolated samples and magnetic-beads isolated samples since those obtained from Melon-gel purification were not of sufficient purity. Our results indicate that magnetic beads purified anti-ssDNA antibodies might have hydrolytic activity against ssDNA. It appears that this activity retained within anti-ssDNA antibody IgG molecule during and after purification.
The anti-DNA antibody isolation and purification methods designed and applied in the past were mainly created to detect anti-DNA antibody structural and functional characteristics (e.g. substrate binding, hydrolytic, and cytotoxic activity).1,7,16,17,21,33 Our two-step affinity purification method was designed to target anti-ssDNA IgG antibodies.19,20 However, our attempts to purify anti-ssDNA antibodies using other oligomers (dC, dG, and dA) bound to streptavidin (SA) coated Dynabeads® were unsuccessful (data not shown). This agrees with results of X-ray crystallography binding studies performed by Tanner et al, (2001, 2004, 2007) which demonstrate that anti-ssDNA mouse monoclonal antibody requires only thymidine pentamer for high affinity binding .This was used as a principal (physical-chemical bases) (Figure 1a) for our magnetic beads design, and has been confirmed to function in the case of human normal, SLE and B-CLL antibodies from plasma.
In summary, Melon-gel (Figure 1b) was far less efficient in purifying anti-DNA ss-DNA antibodies than the magnetic-bead method. Melon Gel isolation of anti-dsDNA antibodies was also performed within our laboratory29 and also showed additional bands, not present after our two-step bead purification (data not shown). As previously mentioned, ssDNA cellulose (Figure 1c) did not work and so was not analyzed further.
The original two-step magnetic bead method designed in our lab gives a reasonable yield (as modified, 40%), is inexpensive, fast (4.5hours) and highly specific, (extracts only ssDNA autoantibodies as visualized by nanoscale sensitive silver staining). These advantages make it a highly useful method for isolation of pure anti-ssDNA autoantibodies. The extensive loss of antibodies during the procedure could be due to numerous wash steps and stirring of the beads during procedure, however, the wash steps are necessary to obtain pure antibody. On the other hand, the yield can be easily increased by increasing the number of beads and the volume of serum used.20 The detection of polyclonal antibodies in lupus sera in comparison to monoclonal antibodies in the case of B-CLL and clinically silent individuals, indicates that the lower yield is not due to possible differences in affinity binding in polyclonal antibodies. This is further supported by the fact that, the highest concentrations of antibodies purified using the two-step method were seen in the lupus serum samples.20 We were not able to determine the recovery rate for the Melon Gel method due to the level of impurities in the final product.
Electrophoretic analysis (Figures 2a & 2b) of purified samples by both methods confirmed that magnetic bead purification is superior with respect to purity and precise antibody concentration determination as shown by Bradford assay (Table 4) (Figure 5).
Western Blot and enzymatic antibody mediated reaction (Figure 4) provided a visual confirmation of the presence of highly purified IgG molecules in the samples obtained via the magnetic bead purification procedure, in comparison to melon-gel. This is the strongest argument for the superiority of our novel magnetic bead method for anti-ssDNA antibody isolation.
The dramatic differences in the electrophoretic patterns (number of bands and MWs) of anti-DNA antibodies isolated from normal, CLL and SLE patients might reflect different mechanisms involved in modulation of autoantibody production, the reasons for which are at this time still unknown (Table 3) Agilent 2100 data versus SDS_PAGE electrophoretic patterns). Hidden patterns identified by the more precise Lab on Chip method (Table 3) confirmed electrophoresis results. Might this mean that clinically silent anti-DNA positive individuals and B-CLL patients have similar B-cell producers? To our knowledge, this type of analysis has not been done before and so we cannot compare our Agilent data to any other information of this type.
The immunoglobulin genes encoding anti-DNA antibody in lupus patients might undergo more somatic mutations and encode more basic amino acid residues than genes encoding natural anti-DNA antibody in healthy subjects and patients with CLL. It is known that the Immunoglobulin gene is composed of VDJ regions on three different chromosomes, (e.g. 2, 14 and 22), and that their rearrangement in autoimmune diseases might be not perfect during polyclonal expansion that takes place during the flare.30 Therefore, somatic mutations are one possible mechanism for explanation of different patterns observed in sera of patients with SLE. Since these antibodies have higher MW (from 165.3-174KDa), as confirmed by Lab-on-chip, two additional epigenetic mechanisms, glycosylation or chain elongation, could also be proposed to contribute to those “patternization,”.19,20 It is more difficult to explain what kind of mechanism could work in B-CLL which is a lymphoproliferative disorder. We do not know the characteristics of the lupus B-cell, however, the patternization of IgGs found in our pilot study might suggests that, at least in this context, B-CLL cells could not be of the same category as anti-DNA antibody producing lupus B-cells.
Different degrees of antibody glycosylation, has been described and determined in autoimmune individuals. Since 1995, Ravetch et al and other groups50,54,55 have published a series of papers indicating that glycosylation of IgGs in rheumatoid arthritis (RA) is one of the crucial underlying molecular mechanisms causing inflammation. IgG antibodies are potent inducers of proinflammatory responses. During autoimmune diseases such as RA and SLE, IgG auto-antibodies are responsible for the chronic inflammation and destruction of healthy tissues by cross-linking Fc receptors on innate immune effector cells.37
The glycosylation of the circulating immunoglobulin - (IgG) antibody molecules changes in rheumatoid arthritis. The extent of the changes correlates with the disease severity and reverses in remission. They demonstrated that the alteration in glycosylation associated with RA can create a new mode for the interaction of IgG with complement through binding to the collagenous lectin mannose-binding protein (MBP).36 Furthermore, RA is associated with a marked increase in IgG glycoforms that lack galactose (referred to as G0 glycoforms) in the Fc region of the molecule and that terminate in N-acetyl glucosamine (GlcNAc). This group has shown using nuclear magnetic resonance (NMR) and X-ray data, that these terminal GlcNAc residues become accessible for MBP binding. They further demonstrated that multiple presentation of IgG-G0 glycoforms to MBP results in activation of complement. This suggests that a contribution to the chronic inflammation of the synovial membrane could arise from the localization of the IgG-G0 glycoforms in the affected joint and from resulting activation of complement. Whether or not something similar also occurs in lupus deserves to be investigated.
Finally, the elongation of the antibody light or heavy chain due to inadequate processing of the protein could be another factor involved in occurrence of banding patterns.37–39 Are these mechanisms operating within lupus B-cells? It is of significant importance to answer those questions since some authors propose that pathogenic anti-DNA antibodies are able to interact with various cell surface proteins (e.g. myosin 1), which enables antibody penetration into the cell.44,45,46 Upon entry into the cell, anti-DNA antibody is translocated into the nucleus where it binds to DNA and consequently hydrolyzes it.
An alternative mechanism by which anti-DNA antibody may lead to auto-immune disease is through its interaction with cell-death receptors, initiating a pro-apoptotic signal that apparently leads to tumor-cell death in vitro and lupus B-cell death in vivo.5,16,17,32 Besides their direct hydrolytic and cytotoxic effects upon the cells observed in vitro, the pathogenic mechanisms involved in SLE include the induction of initial lesions via deposition of circulating immune complexes (composed of lupus DNA and antibodies bound to it) in the tissues of various organs in vivo. Deposition in the tissues induces an inflammatory response or cytotoxicity, and leads to multiple organ/tissue damage in the form of associated disorders such as systemic vasculitis, glomerulonephritis, chorea and others. The most important pathological finding in nephritogenic lupus is the presence of immune deposits beneath the glomerular endothelium in kidney biopsies and virus-like particles in endothelial cells.48 The presence of virus-like particles in different organs in patients with lupus indicate that the anti-DNA antibodies could be a part of the body’s viral or bacterial defense mechanism involved in the removal of foreign DNA during the course of the disease.49
In summary, we could conclude that the reason for different structural patterns of anti-ssDNA autoantibodies in SLE could be the product of genetic and epigenetic phenomena. Proposed scenario: during disease some genes are turned on to produce somatically mutated antibodies, while epigenetic phenomena such as glycosylation and elongation make them potentially more or less inflammatory and hydrolyticaly active. Only further investigation can confirm or dispute this postulate.
The oscillations in ELISA results for anti-ssDNA antibody concentration determination (Table 5) (Figure 6) have shown that system is not suitable for our purposes. Originally, oligo-dT coated plates were used in our lab19 with excellent quantitative results using a plate reader with 4-logistic log system.27 It has been reported that in most lupus patients and normal donors IgG1 and IgG4 are the most abundant (>30%), while IgG2 and IgG3 are present at lower concentrations (<30%) (1). Proportions of the four human IgG subclasses, as a survey is provided in Table 6. Results of ELISA for IgG subclasses (Figures 7-9) (Table 6) have shown that there is similar distribution of all 4 subclasses obtained when both methods are tested, with the exception of IgG4 which is much higher in Melon-gel treated samples. According to the literature, hydrolytic anti-DNA antibodies are reported to be of IgG1 and IgG3 subclasses.1,8 Therefore, the possibility whether or not anti-DNA antibodies of all of these isolated subclasses express hydrolytic/cytotoxic activity should be examined.
The results of isoelectric focusing IEF, (Figure 10a & 10b) also corroborate that the magnetic bead method reveals the polyclonal nature of purified human lupus antibody (a smeared pattern, pI~9) compared to mouse monoclonal antibody (three clearly distinctive bands pI~7). These data suggest that the purified antibodies from the lupus patients are polyclonal and that the B-CLL and normal donor antibodies were monoclonal, thus accounting for the different banding patterns. This is also an indication that B-cells in B-CLL and normal individuals are closer in evolution than lupus B-cells.
UV and Lab-on-chip results, (Figures 11 &12) have also demonstrated that anti-ssDNA antibody could have hydrolytic activity, as some authors have already detected in the case of mouse monoclonal anti-ssDNA antibody using Gololobov’s modified sequences as a substrate and analyzing results of electrophoresis.34 They have shown that this antibody was capable of hydrolysis in its IgG and IgM modalities and that the activity of IgM was 17 times higher than IgG.34 The authors have also examined the activity of purified anti-IgG Fab fragment, after papain hydrolysis and found that its activity is three times higher than the activity of the entire intact antibody molecule. They stressed the possibility that the Fc fragment, eliminated in papain hydrolysis, could have inhibitory effects upon this reaction.7,21,34 Our results, to our knowledge for the first time, confirm the hydrolytic activity of pure, polyclonal, human anti-ssDNA lupus antibody. Future work is necessary to increase the sensitivity levels of the hydrolytic assays, locate the specific position of the hydrolytic component of the antibody and determine its reaction kinetics in comparison to DNAse1 in order to prove the antibody’s intrinsic hydrolytic capacity. Armed with all of these tools we believe that it will be possible to prove its pathogenicity through further examination.
The distinction between anti-ds and anti-ssDNA auto-antibodies is not quite clear (Table 1) and additional research will be required to explain the presence and reason for the presence of anti-ssDNA auto-antibodies in 30-70% of patients with autoimmune diseases including SLE.33,34 There is a lot of controversy surrounding the potential immunogenicity of mammalian (human) DNA. One of the most recent reports of note demonstrates the detection of ssDNA antigen in human serum from patients with SLE by microarray.51 Most researchers state that by itself, it is not immunogenic, due to nucleotide monotony, but becomes more intriguing to the immune system with ageing and unmethylation. If self-ssDNA could not be considered immunogenic until some later period of life, what is the ssDNA immunogen in childhood SLE?
Other hypotheses implicate viral DNA,7,18,47,50 microbial DNA, and/or unmethylated CpG motifs within the DNA of any species56 all of which may be interesting to the innate immune system. Could a new mechanism of antigen recognition be taking place, omitting the classical antigen-presentation loop to the T-cells and their helper response? DNA-sensing proteins in inflammasomes, could convey signals to TLR9 and through MyD88, activate the transcription factor NF-kappa B which is responsible for antibody production in germinative B-cells.
The origin of anti-DNA antibodies and the antigenic impact of DNA source in SLE and other autoimmune diseases is still a matter of intensive debate.7,18,47,50 Furthermore, all of the same questions that we have about anti-ssDNA antibodies can also be applied to the dsDNA category.
In this analytical study, compared to the magnetic bead method, Melon gel allowed for faster antibody purification and higher recovery rate, but produced a final product of significantly lower purity when analyzed by both PhastGelTM and Agilent 2100 methods. Electrophoreses and Lab-on chip analyses have revealed that purified anti-DNA antibodies from the sera of patients with SLE express electrophoretic patterns that differ from normal controls and vary between individual SLE patients. The meaning of this “patternization” is unknown, but could be due to somatic mutation, glycosylation and/or elongation. IEF data strongly suggested polyclonal nature of purified anti-DNA antibodies, in SLE and monoclonal in normal donors and B-CLL patients.
ELISA results revealed that anti-DNA antibodies from SLE patients belong to all 4 subclasses with different participation. This is contrary to the generally accepted belief by researchers and clinicians that IgG1 and IgG4 are the most abundant iso-types.
The excellent sensitivity, reproducibility and convenience in combination with specific and fast antibody purification via the two-step magnetic beads method will enable this method to be widely useful for the study of both double and single-stranded DNA cleavage reactions important for cellular events.
Therefore, it can provide a deeper insight into the pathogenicity of anti-DNA antibodies in lupus disease and discriminate between the roles of anti-ssDNA versus anti-dsDNA antibodies. Simple modification of this method will also allow for comparison of anti-DNA IgG and anti-DNA IgM hydrolytic activities, since according literature data, both isotypes have shown to be hydrolytic.6,7,45 Analysis for antibody hydrolytic activity against a herring sperm ssDNA substrate, via UV spectrophotometry and Lab-on-chip data, have shown that lupus patient antibody isolated via magnetic beads has different reaction kinetics than DNA-se1 indicating that hydrolysis is intrinsic property of the antibody molecule.
In summary, we conclude that application of our two step purification method would be of fundamental importance for lupus diagnosis, flare prediction, monitoring of the disease, and preventive therapy, and will give an insight into molecular mechanisms of its pathogenesis.
Phyllis and Dudley Whitman who have supported this work through Florida Atlantic University Lupus Foundation. We also acknowledge and deeply appreciate the proof reading and critical remarks of Ms. Michelle Cavallo and Dr JamesxHartmann.
The author declares no conflict of interest.

©2017 Kats, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.