MOJ
eISSN: 2374-6920


Research Article Volume 5 Issue 1
1Department of Biotechnology, Guru Nanak Dev University, India
2Centre of Systems Biology and Bioinformatics, Panjab University, India
Correspondence: Ashok Kumar, Centre for Systems Biology and Bioinformatics, Block-III, Sector-25, Panjab University, Chandigarh-160014, India
Co-correspondence: Ramandeep Kaur, Department of Biotechnology, Guru Nanak Dev University, Amritsar-143005, India
Received: October 05, 2016 | Published: January 26, 2017
Citation: Singh SK, Patiyal S, Kaur R, et al. Prediction of metal ion binding site in truncated globin of Myxococcus xanthus DK 1622 in homologous model. MOJ Proteomics Bioinform. 2017;5(1):7-12. DOI: 10.15406/mojpb.2017.05.00146
Globins are heme containing proteins with the capability to bind oxygen reversibly. All globins can be classified as M (myoglobin-like), S (sensor) and T (truncated) globins; among them, M and S globins have a typical 3/3 α-helical globin fold and T globins have a 2/2 α-helical fold. Till date, all the myxobacterial genomes available in NCBI data base harbor one or more genes for globins but none of the globins from this group of bacteria have been characterized as yet. In this study, in-silico analysis and homology modeling approach was exploited to explore the physiochemical properties and 3-D structure of trHbN (group I T-globin) of Myxococcus xanthus DK1622, a fruiting myxobacteria. After generating the 3-D model, assessment of the structure and stereo-chemical analysis was carried out using different evaluation and validation tools. As homologs of the trHbN exist in dimeric as well as monomeric form, dimerization studies were performed and it was observed that dimeric form of the protein is more stable as compared to the monomer. Further, heme group was appended to the model structure of trHbN of M. xanthus by Patchdock web server and model-heme complex was further refined by Firedock. The 3-D model structure will enhance the understanding for functional analysis of experimentally derived crystal structure.
Hbs, hemoglobins; GCS, globin coupled sensor; SDSGb, single domain sensor globin; NO, nitric oxide
he presence of hemoglobins (Hbs) has been reported in all forms of life; from simple prokaryotes to complex eukaryotes; and share a common characteristic of reversible oxygen binding.1 These globins play a role in diverse functions, such as, oxygen transport, storage of oxygen, terminal oxidases, oxygen sensors and are also involved in response to oxidative and nitrosative stress.2–6 The structures of vertebrate myoglobin as well as hemoglobin have a typical tertiary structure consisting of 8 alpha helices (A-H) arranged in 3 over 3 topology connected by short intervening loops.1 All the globins are classified into three distinct lineages; the M (myoglobin like) family, the S (sensor) family, and T (truncated) family.7,8 Flavohemoglobin (FHb) and single domain globin (SDgb) constitute the M globin family; the S family comprises of globin coupled sensor (GCS), single domain sensor globin (SDSgb) and protoglobin9,10 (Figure 1). T globins are usually 20-40 residues shorter than the vertebrate Hbs and display 2 over 2 helical structures. T-globins are further classified into group I, II and III designated as trHb1, trHb2, and trHb3 respectively.3,11,12 In all the globins, His-F8 is the strictly conserved residue that forms co-ordinate bond with the iron of the heme.
There are limited reports on the in vivo role of truncated globins and these studies suggest that group I 2/2HbNs are associated with nitric oxide (NO) detoxification and the group II 2/2HbOs are involved in oxygen metabolism. Till date, the function of the group III 2/2HbP has been characterized only from Campylobacter jejuni and its role is not very clears The T-globins from more than one group are found to coexist in an organism indicating the functional divergence of these globins.
Myxobacteria are unicellular, rod shaped Gram-negative bacteria that belong to the delta class of proteobacteria. They predominantly inhabit terrestrial but their presence is also reported in aquatic environment and they have been isolated from tree bark and animal dung. The genomes of the Myxobacteria have high GC content of ~70%.13,14 Myxobacteria have unique characteristics among the bacteria as they show coordinated motility, predation and produce multicellular fruiting bodies under nutrient limited condition.15 These bacteria produce several bioactive secondary metabolites of pharmaceutical importance and have contributed significantly to the human health.16 The genomes of the Myxobacteria contain genes for globins from all the three lineages.17 It is possible that the presence of different globins in a genome of myxobacterium is correlated with the diverse functions of the globins in different stage of life cycle.
Myxococcus xanthus DK1622 belongs to the order myxococcales and is a strict aerobe. The genes for two truncated globins (T globin group I and II) and a globin coupled sensor are present in the genome of M. xanthus DK1622.17 As the globins from myxobacteria are neither characterized nor their experimental 3D structure is available, the aim of the present study was to characterize the physiochemical properties of trHbN from M. xanthus DK 1622 by in-silico approaches and comparative modeling of the trHbN with the special reference to the prediction of metal ion binding site and its biological conformation. Therefore, this study provides an insight into the structural features of trHbN from M. xanthus DK1622 to understand the ligand binding characteristics of this globin.
Amino acid sequence
The protein sequence of the trHbN (Accession number: WP_011556840.1) of Myxococcus xanthus DK1622was retrieved from NCBI protein sequence database in FASTA format and used for further analyses.18–20
Physico-chemical characterization
Physicochemical parameters like Iso-electric point, extinction coefficient,20 amino acid composition, positively charged residues; negatively charged residues, aliphatic index,21 instability index22 and GRAVY23 of the trHbN of myxobacteria were analyzed using Expasy’s ProtParam web server.24
Secondary structure prediction
Secondary structural analysis of trHbN was done using PSIPRED v3.3 web server.25
Molecular modeling
PSI-BLAST search against the Protein Data Bank using protein sequence of the trHbN from M. xanthus DK1622 was performed and best template with good sequence identity (>30%) and lowest E value was selected for homology modeling. Template selection was further refined by using template selection option from SWISS-MODEL web server.26
The assessment of the template was tested at every step to provide the best template for homology modeling. Primary amino acid sequence was compared using web tool EMBOSS Needle27 to check the identity and similarity between the query and the template. Secondary structure predicted by PsiPred v3.3 was compared to find out the similarity in the secondary structure pattern (helix and coil) between the selected template and the query. Finally, 3-D model of trHbN was generated by Modeller28 and the predicted 3D structure was validated for the structural and stereochemical quality by using Varify3D29 and ERRAT30 from SAVES web server, PROSA web31 and PROCHECK.32 Interactive visualization and analysis of molecular structures was carried out in UCSF Chimera.33
Homodimer generation
Functional form of the homologues of the experimentally proved trHbN from other microorganisms was found to exist as dimer as well as monomer; therefore, homodimer was generated using online server for protein-protein docking ClusPro2.0.34 The PDB file of the best predicted 3D model of the trHbN of Myxococcus xanthus DK1622, was provided as the input for dimerization. The dimerization was done using multimer docking plugin in the advanced option of cluspro 2.0 and the cluster of dimers with various orientations were retrieved and the energy for each of them was calculated and analyzed.
Stability analysis of dimer of the predicted trHbN
The stability of the dimeric structure was determined by calculating the free energy of the monomer and the dimer using GAFF force field in the standalone software Avogadro 1.2.0. The free energy of the protein structures was calculated with and without optimization using GAFF force field for monomer and dimer.
Molecular docking
The docking analysis of heme was carried out with the online server for docking -PatchDock v beta 1.3.35 SDF model file of heme was retrieved from the Protein Data Bank (PDB) and converted in to PDB file in USCF Chimrea. The PDB files for the receptor (3D model of the trHbN of M. xanthus) and ligand (heme) uploaded in the PatchDock web-server with clustering RMSD value 1.5 and protein-small ligand as complex type were selected as parameter for docking. The structure-ligand complex generated by PatchDock was further refined by Firedock36 on the basis of free energy. 2D image of heme and protein interaction was plotted with LIGPLOT+.
Physicochemical characterization
Amino acid sequence plays a very important role in determination of 3D structure of any protein, as each amino acid has a different propensity to form secondary structure like helix, beta-strand, coil and turn. Therefore, it is necessary to characterize the physicochemical parameters of a protein before analyzing its 3D-structure. In the present study, Expasy Protparam web server was used to characterize the physicochemical parameters of trHbN of M. xanthus. The amino acid sequence of the hypothetical protein was predicted to have 126 amino acids with molecular weight of 13579.53 Da and theoretical isoelectric point (PI) of 6.04. Extinction coefficient was calculated to be 4470 (with 10% error in the absence of tryptophan residue). The instability index (II) was calculated to be 36.13 and classified as the stable protein. The negative Grand average of hydropathicity (GRAVY) of -0.055 indicates that the protein is hydrophilic and soluble in nature. Alanine (15.9%), leucine (7.9%), glutamic acid (7.10%), histidine (4.8%), and methionine (4.8%) constitute the major fraction of amino acids of the trHbN and have greater propensity to form helical structure.
Secondary structure content
The PSIPRED v3.3 results revealed that only helices and coil are secondary structure elements found in amino acid sequences of the trHbN of M. xanthus DK1622 (Figure 2).
Structure of trHbN of xanthus DK1622
No experimentally determined structure of the group I 2/2 HbN of M. xanthus is available till date. Therefore, we built a 3D-model following a homology modeling protocol.
Homology modeling and quality assessment: A PSI-BLAST search against PDB database was performed to find suitable templates for homology modelling and template selection was further refined by SWISS-MODEL template selection option. Based on the maximum identity (with high score) and lower e-value, crystal structure of trHbN of Chlamydomonas reinhardtii (PDB ID: 4XDI_A) at 1.9Å resolution37 was selected as the best available template to build a 3-D model of trHbN of xanthus DK1622 with MODELLER9.15. The target sequence displayed 44.5% identity and 62.5% similarity with 4XDI_A used as templates in pairwise sequence alignment performed using EMBOSS Needle web server (Figure 3). Root mean square deviation (RMSD) value between models (generated by MODELLER) and template (4XDI_A) was calculated by structural superimpositions of Cα pairs of the predicted structures with the template (UCSF Chimera, interatomic distance threshold -2Å). The predicted model structure having the least RMSD (0.227Å) value with the template’s structure was selected as the putative 3D structure for further analysis.
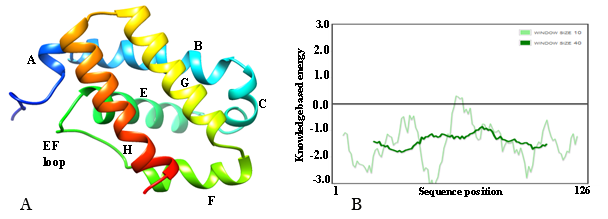
The quality of final 3D model of trHbN was assessed by different web tools and invariably indicates good quality of the model. Structure analysis by VERIFY 3D revealed that 96.03% of the residues had an average 3D-1D score >= 0.2, indicating a valid model. The VERIFY 3D program was used to determine the compatibility of an atomic model (3D) with its own amino acid sequence (1D) by assigning a structural class on the basis of its location and environment (alpha, beta, loop, polar, non-polar, etc.) as well as comparing the results with good database structures.29
Overall ERRAT score (quality factor value), expressed as the percentage of the protein for which the calculated value falls below the 95% rejection limit was 98.305%. Good high resolution structures generally produce values around 95% or higher.30
A global and local stereochemical property on the basis of interaction energy per residue of the structure was interrogated with a Z-score (PROSA web server). Z-score of the 3D structure calculated from PROSA Web server was found to be -6.4 which indicates reliable model. Finally, the stereochemical excellence as well as overall structural geometry of the predicted 3D structure of trHbN was confirmed by PROCHECK at minimum 2.0 Å resolutions. Ramachandran plot drawn through PROCHECK program validated the model with 97.3% and 2.3% in the most favored and additional allowed region respectively. No residue was found in the generously allowed as well as disallowed region. Total quality of goodness (G) factor value of the model was calculated to be 0.09, which is in the range of the acceptable good quality model (0 to -0.5). These results revealed that the distribution of the protein backbone dihedral angles (phi and psi) occupied reasonably accurate positions in the 3-D model.
These results ensure that the model was predicted with good accuracy and can be used as a template for further investigation.
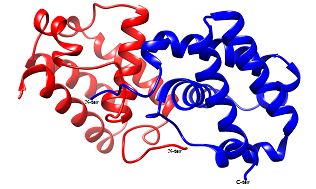
Prediction of dimer assembly of modeled structure: Functional forms of the homologues of the experimentally proved trHbN from other microorganisms were found to exist as dimer as well as monomer. Therefore, to get insight into the oligomeric state of the trHbN, computational algorithm was employed to predict the functional form of the trHbN. Homodimer of the predicted structure was generated using automated docking web server ClusPro 2.0. The server generated 14 clusters of dimer conformation, out of which cluster-0 with lowest energy (-728.9KJ/mol) was selected and all the dimer conformations from cluster-0 were considered for further analysis.
Force field analysis: Stability of the predicted model depends on the energy of the structure i.e. lower the energy more stable is the structure. Therefore, energy of the dimer and monomer was calculated using the GAFF force field in the standalone software Avogadro 1.2.0 and on comparison it was found that the dimer has lower energy compared to monomeric form, indicating that dimer might be the preferred stable structure of the trHbN in the native host. The energies were calculated for each structure and compared in Table 1 & 2.
Biological assembly |
Energy (Kj/Mol) |
Minimized energy (Kj/Mol) |
|
Chlamydomonas reinhardtii |
Monomer |
6,425.76 |
-414.636 |
Dimer |
2.69 x 1020 |
361.321 |
|
Myxococcus xanthus |
Monomer |
6,913.04 |
-1,409.15 |
Dimer |
5,330.54 |
-3,230.02 |
|
Table 1 Comparative force field analysis of Globins
Name of the structure |
Metal ion co-ordination geometry tables |
|||
Co-ordinator |
Geometry |
Co-ordination number |
RMSD value |
|
Predicted trHbN of M. xanthus |
His-F8 (75) |
Octahedron |
6 |
0.266 |
Lys-E10 (51) |
Octahedron |
6 |
0.322 |
|
trHb1 of C. reinhardtii (4XDI_A) |
His-F8 (77) |
Octahedron |
6 |
0.054 |
Lys-E10 (53) |
Octahedron |
6 |
0.072 |
|
Table 2 Comparison of metal ion geometry of the trHbN of M. xanthus with crystal structure of trHb1 of C. reinhardtii
Docking of heme group in trHbN: Docking studies explore the new path to build up the functional structure of the 3D model of the protein and crucial information regarding the residues involved in protein ligand interaction can be derived. Heme is an iron containing cofactor required to hold the diatomic gaseous ligand to fulfill the function of the globins. Therefore, docking of heme in the predicted model of the trHbN is very crucial step to illustrate the structure and functional relationship of the hypothetical putative protein trHbN. Patch dock performs rigid docking and molecular docking algorithm is based on the principle of shape complementarity. Docked structure generated and refined by Patchdock and Firedock respectively were analyzed using the stand alone visualization software USCF Chimera 1.10.
Docking of heme in the 3-D model of the trHbN of M. xanthus predicted some of the key residues that are involved in interaction with the heme moiety. From the structure alignment of the template (4XDI_A) and predicted 3-D model docked with heme, it appears that histidine-75 and lysine-51 are involved in interaction with iron of the heme, similar to the histidine-77 and lysine-53 of the trHb1 of C. reinhardtii. Crystal structure of the trHb1 (4XDI) of C. reinhardtii used as template in this study, was solved at ferric state which demonstrates distinct features associated with the coordination of the heme iron by histidine at proximal side and lysine at distal side.37 Therefore; from the above prediction it can be concluded that trHbN of M. xanthus may be hexacoordinated with 2 proteinaceous ligand, histidine-75 and lysine-51 at proximal and distal side of the heme respectively.
The metal ion geometry for the predicted model as well as trHb1 of C. rheinhardtii was compared using UCSF Chimera 1.10. It was found that the residues interacting with the iron of heme in predicted model of the trHbN of M. xanthus DK1622 are His-75 and Lys-51 with octahedron geometry and coordination number as six; similar geometry was found in the crystallized structure of THB1 of C. reinhardtii with residues His-77 and Lys-53 (Figure 4) (Figure 5). This further supports the conclusion that trHbN of M. xanthus may be hexacoordinated with 2 proteinaceous ligand, histidine-75 and lysine-51.
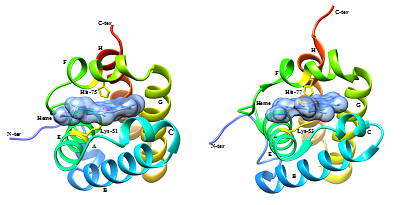
Other residues that may be involved in interaction with heme are phenylalanine-40 (CD1), phenylalanine-55 (E14) and tyrosine-66 (EF6). It was observed that Arginine-47, present in trHbN of M. xanthus in place of lysine-49 of 4XDI, interacts with one of the propionate heads of the heme. Lysine-69 interacts with second heme propionate head through hydrogen bond whereas in the trHb1 of C. rheinhardtii Arginine- 52 interacts with this propionate head. Heme is embedded in the hydrophobic region of the protein known as heme binding pocket, which is composed of hydrophobic amino acids to protect the solubility of heme and these hydrophobic amino acids stabilize heme through vanderwaals interactions in all 2/2HbNs as shown in ligplot (Figure 6).

These findings are potentially important to be used for further annotation of the globins of the T- family from myxobacteria and can be used to link the specific functions to their respective structures. The results presented here seem to be relevant as we show (i) the dimer form of the globin is much stable as compared to the monomer form, which also could be its functional form and (ii) a molecular model showing interaction between iron of heme and histidine residue of the group 1 truncated globin, which exhibits the conservational interaction in the T family of globins. These facts can be used in future for further classification or functional analysis of the various classes of globins in the myxobacteria.
To enhance the insight in structure-function relationship of the trHbN of M. xanthus DK1622, several computational web tools were used to characterize the physiochemical properties, secondary structure prediction as well as generation of 3-D structure of protein by homology modeling. Structural and stereochemical quality of the predicted model was evaluated with Varify3D, ERRAT, PROSA and PROCHECK. Outcome of these tools invariably indicates good quality of predicted 3-D structure. Further biological assembly of the trHbN was generated using Cluspro2.0, an automated tool for protein-protein interaction and dimer was found favorable in terms of free energy.At the end, docking of the heme group in trHbN indicates histidine-75 and lysine-51 as apparent metal ion binding site for the heme moiety through co-ordinate bond on the proximal and distal side of the heme respectively, while other residues are involve in interaction with heme through hydrogen bonding or hydrophobic interaction. Outcome of the study can be used to investigate structure function relationship of other globins in myxobacteria.
Authors are thankful to Department of Biotechnology (DBT), Government of India, New Delhi, India to provide the funds necessary for financial assistance in the form of DBT-JRF programme to one of the author SKS [DBT-JRF/10-11/242, dated: 31-08-2010].
The author declares no conflict of interest.

©2017 Singh, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.