MOJ
eISSN: 2374-6920


Research Article Volume 5 Issue 1
Department of Urology and The Proteomics Center, Boston Children's Hospital and Harvard Medical School, USA
Correspondence: Richard S. Lee, Department of Urology and The Proteomics Center, Boston Children's Hospital and Harvard Medical School, USA, Tel 6173553348, Fax 6173557796
Received: November 16, 2016 | Published: January 20, 2017
Citation: Zhou H, Warren PG, Froehlich JW, et al. 18 O-incorporated de-n-glycosylated site identification: a comparison of three search engines. MOJ Proteomics Bioinform. 2017;5(1):1-5. DOI: 10.15406/mojpb.2017.05.00145
Enzymatic de-N-glycosylation has become an essential procedure in most large scale studies to identify N-glycosylation sites because the removal of heterogeneous sugar moieties enables the detection of deglycosylated peptides by standard bottom-up proteomic pipelines. To differentiate deglycosylation from chemical deamidation, this reaction is often carried out in an H218O environment, resulting in a conversion of Asn to 18O-Asp (+2.9883Da). The detection of the alteration is dependent on the use of available search engines. We performed a large-scale comparison of three commercial search engines, ProteinPilot, Mascot, and Sequest, to assess their capability to identify the 18O-incorporated aspartic acid. To compare the results from the three search engines, peptides obtained from three different urine samples were separately filtered by each search engine at a 1% peptide false discovery rate and proteins were identified with a minimum of 2 distinct peptides. Of the three search engines, ProteinPilot identified the largest number of proteins, peptides, non-modified consensus motifs and deamidated motifs. However, both Sequest and Mascot detected more unique 18O-incoporated motifs than ProteinPilot. Interestingly, the majority of proteins identified with a unique 18O-incoporated motif by both Sequest and Mascot were also identified by ProteinPilot. Overall, ProteinPilot demonstrated a lower sensitivity to specifically identify 18O-incorporated motifs, but not glycoproteins that contained those motifs. Our results may help improve future large-scale N-glycosylation site identification studies.
Keywords: mascot, protein pilot, Sequest, pngase f, deglycosylation, deamidation, 18o-incorporated asp
As one of the most abundant post-translational modifications of mammalian proteins, N-glycosylation takes part in nearly all physiological and pathological activities.1 Furthermore, N-glycoproteins have become focused targets in numerous large scale biomarker studies because N-glycosylated proteins are often enriched in body fluids such as plasma and urine.2 In fact, the majority of clinically-used protein biomarkers are glycosylated.3 Various strategies have been employed to characterize glycosylation and glycoproteins in biological samples.4 Most mass spectrometry-based studies typically study one of three distinct analytes: released glycans (also referred as glycome for complex samples),4 deglycosylated peptides (deglycopeptide) in which sugar residues are completely removed from the modified asparagine,2 and glycopeptides in which glycans are still attached to the peptide.5,6 Interrogation of deglycopeptides will likely remain the most attractive analyte due to numerous analytical advantages. First, the removal of complex and heterogeneous sugar residues enables the detection, characterization, and even quantification of deglycopeptides by standard proteomic methodologies.7 Second, the stripping off of sugar-moieties allows the detection of both deglycopeptides and their counterpart unmodified (non-glycosylated) peptides simultaneously, which are necessary to identify partially glycosylated sites unambiguously.8 Third, the sugar residues can be employed to enrich for glycoproteins or glycopeptides using hydrazide-chemistry or lectins.7,9
In a typical N-glycosylation site profiling study PNGase F is utilized to detach N-glycans from the asparagine within the N-glycosylation consensus motif (Asn-X-Ser/Thr, where X is any amino acid except proline),10 resulting in the conversion of the sugar-attached asparagine to an aspartic acid (Asn to Asp, +0.9840Da). Unfortunately, this product is identical to those derived from spontaneous chemical deamidation,11 which may occur in vivo or in vitro. Researchers have shown that performing the enzymatic reaction in an H218O environment is effective in differentiating enzymatic conversion (Asn to 18O-Asp, +2.9883Da) from chemical deamidation.8,9 Identifying 18O-incorporated deglycopeptides usually involves searching MS/MS spectra against an appropriate protein Database using various search engines such as Mascot,12 ProteinPilot,13 and Sequest.14 Such characterization fully relies on the capability of employed engines to identify 18O-incorporated aspartic acid residue, which are the basis for site identifications or peptide quantifications.2 So far, there has not been a dedicated study evaluating the capabilities of different search engines with respect to identifying 18O-incorporated deglycosites (formerly-occupied glycosylated sites). Herein, we systematically compared the performance of three common search engines (Mascot, Sequest, and ProteinPilot) in their ability to detect 18O-incorporated deglycosites in human urine specimens.
Materials and reagents
PNGase F (glycerol free) was obtained from New England Biolab. (Ipswich, MA). Sequencing grade trypsin was purchased from Promega (Madison, WI). The Viva Spin 2 series of spin filters (30K MWCO, 2mL volume, Polyethersulfone-type membrane) were purchased from Sartorius Stedium Biotech (Aubagne, France). H218O (98%) was obtained from Rotem (Arava, Israel). All other chemicals and reagents, if not specified, were purchased from Sigma-Aldrich (St. Louis, MO). Centrifugation was performed in a fixed-angle rotor of a bench-top centrifuge 5804R (Eppendorf). The default spinning period was 20min at 10,000g, unless otherwise specified.
Urine processing
The urine specimens were obtained from three healthy donors under an institutional review board-approved protocol. The depletion of albumin in urine was performed according to a published one-step protocol,15 and the protein concentration was measured by the Bradford assay in triplicate. Approximately 250µg of depleted urinary proteins from each donor were further processed according to the GlycoFilter platform to obtain released N-glycans and tryptic peptides,8 respectively. PNGase F catalyzed de-N-glycosylation was carried out in a H218O buffer via a 20-min domestic microwave protocol.16
Tryptic peptides from three different urine samples were focused into 24 fractions using a 3100 OFFGEL fractionator (Agilent, Santa Clara, CA) as described previously.15 Briefly, the 24cm, pH 3-10 IPG DryStrips (GE healthcare) were rehydrated for 20min with the IPG buffer pH 3-10. Samples were dissolved in 3.6mL of IPG buffer, and equally distributed into each well. Focusing was performed according to the preset program up to 50kV h with maximum current of 50mA. Fractions were collected from each well. An additional 100mL of 0.1% formic acid was added to each well, and extracted after 10min. The extracted peptides were combined and dried completely in a speed-vacuum. The fractionated peptides were further desalted with Strong Cation Toptips (Poly Sulfoethyl A, Catalog #TT2SSA) according to the vendor’s instruction (Glygen, Columbia, MD).
LC-MS/MS analyses of peptides
The desalted peptides were analyzed by an LTQ-Orbitrap XL mass spectrometer (Thermo Scientific, Waltham, MA) connected to an autosampler and nanoflow HPLC pump (Eksigent, Dublin, CA). The reversed phase columns were packed in-house using Magic C18 particles (3mm, 200Å; Michrom Bioresource), and PicoTip Emitters (New Objective). The peptides were eluted with a 60minute linear gradient (0-35% acetonitrile with 0.2% formic acid), and Data acquired in a Data dependent mode, fragmenting the seven most abundant peaks by CID, with dynamic exclusion for 60s. All precursor scans were performed in the Orbitrap, and MS/MS spectra were obtained from the low resolution linear ion trap. Buffer A was 0.2% formic acid, buffer B was acetonitrile and 0.2% formic acid, and loading buffer was 5% formic acid with 5% acetonitrile.
Database searching with three engines
ProteinPilot: The 200 most intense fragment ions of each raw product ion spectrum were used to generate. MGF files using the peak-list-generating software ProteoWizard (2.2.2881, released on 2012-7-25). The MGF files were searched by ProteinPilot (V4.5.1) against the UniProtKB/Swiss-Prot target Database (Homo sapiens, released in 2012_07) containing 20,283 protein sequences. The detailed paragon parameters included: sample type, identification; cysteine alkylation, iodoacetamide; digestion, trypsin; instrument, MS1 Orbi/FT and MS/MS LTQ; Special factors, PNGase F in H218O; Species, none; Search effort, thorough ID; Results quality, run false discovery rate (FDR) analysis and detected protein threshold (0.05). Notably, only the target Database was loaded onto the ProteinPilot and Proteome Discoverer (used for both Mascot and Sequest searches); the complete decoy (reverse) sequences were generated by both platforms in situ. The score level equivalent to 1% FDR level (as determined from the respective FDR analysis spreadsheet output) was determined for each sample. This cutoff score was then applied to yield a subset of peptides filtered to 1% FDR.
Sequest and mascot: Thermo raw spectra were loaded into the Proteome Discoverer (v1.3). Sequest (embedded in the Proteome Discover) and Mascot (v 2.3)12 searches were respectively performed within Proteome Discoverer platform against the same UniProtKB/Swiss-Prot human target Database used for ProteinPilot. One missed cleavage per peptide was allowed and mass tolerances were 10 ppm for precursor and 0.8Da for MS/MS fragment ions. The search included fixed modification of carbamidomethylation on cysteine, and variable modifications: oxidation (Met), chemical deamidation (16O, Asn and Gln), and enzymatic de-N-glycosylation (18O, Asn). The upper FDR limit of peptide identification was set at 1% using the percolator module within the Proteome Discoverer.
Protein identifications: Proteins were declared with a 2-peptideminimum. Peptide-to-protein grouping was performed using graphical analysis to regroup all peptide-protein assignments across the three search engines’ outputs, then choosing the same representative protein for any constituent peptides within each search engine. A non-redundant protein Database was used, and peptides which were shared on a protein isoform level were combined into the longest isoform of that protein group. Any remaining shared peptides were removed from consideration. All comparisons were based on a 1% spectral FDR as assigned by individual search engine results.
Deglycosite and glycoprotein assignments: In this study, a deglycosite was defined by satisfying two required criteria: 1) the identified peptide sequence contained the common N-glycosylation consensus motif (Asn-X-Ser/Thr, where X is any amino acid except proline);17 and 2) the asparagine residue within that motif was identified as the 18O-incorporated deamidation derivative (18O-Asp+2.9983Da).9 Glycoproteins were defined as proteins containing at least one identified deglycosite. An internally created R package was developed to identify and count distinct N-glycosylation consensus motifs. The code checked for all identified consensus motifs in any peptide sequence, as well as those spanning the C-terminus of peptides (Asn-Arg-Ser/Thr or Asn-Lys-Ser/Thr for tryptic peptides). It then checked these against the FASTA Database, assigning a specific protein location (amino acid number) to each motif, and removed all redundancy caused by multiple identifications of the same motif (such as those caused by missed cleavages). This ensures that each distinct motif was counted only once, regardless of how many different cleavage forms of peptides contained that motif. The code then determines the identified form of each motif: deglycosylated (18O-Asp), deamidated (16O-Asp), and unmodified (Asn).
In contrast to other sugar-based enrichment strategies, no enrichment was involved in this study, allowing concurrent characterization of all tryptic peptides. The rationale for this approach was to enhance the identification of glycoproteins, by not relying solely on deglycopeptides as in most studies involving enrichment.2 Most importantly, the characterization of unmodified peptides allowed a thorough comparison of all three engines with respect to the identification of all three forms of any specific N-glycosylation consensus motif: unmodified (Asn), deamidated (16O-Asp), and deglycosylated (18O-Asp).
The proteome and glycoproteome comparisons
The identified proteins and peptides, as well as deglycosylated proteins and glycosites from three urine samples via three different search engines are listed in Table 1. In general, ProteinPilot yielded the largest number of non-redundant peptides among all three engines, leading to the identification of more unique proteins. As for Sequest and Mascot, it seemed that Sequest was slightly more sensitive than Mascot in this regard, as Sequest generated more numbers in terms of both characterized proteins and peptides. Since it is well known that the three engines employ different algorithms and scoring systems to characterize peptides, these kinds of discrepancies with respect to proteomic identifications were not unexpected.18 Therefore, we focused our investigation on the identification of deglycosylated proteins and deglycosites. Notably, ProteinPilot yielded the fewest 18O-incorporated deglycosites and deglycosylated proteins (Table 1), as well as fewer deglycopeptides (data not shown) despite identifying the highest numbers of proteins and peptides. In contrast, Sequest identified approximately 20% more deglycosylated proteins than those identified by ProteinPilot (Table 1). Mascot also performed better than ProteinPilot. The difference in identification of deglycosylated proteins between Sequest and Mascot were much smaller (Table 1). These results demonstrate that both Sequest and Mascot appear to perform better than ProteinPilot in their ability to identify deglycosites (18O-Asp).
Sample |
Proteins |
Peptides |
Deglycosylated Proteins |
De-glycosites |
||||||||
SEQ |
MAS |
PP |
SEQ |
MAS |
PP |
SEQ |
MAS |
PP |
SEQ |
MAS |
PP |
|
U1 |
2093 |
1946 |
2321 |
13312 |
12563 |
21299 |
540 |
519 |
426 |
990 |
948 |
757 |
U2 |
1944 |
1797 |
2251 |
11760 |
10984 |
18475 |
503 |
472 |
389 |
887 |
839 |
673 |
U3 |
2028 |
1884 |
2024 |
11968 |
11408 |
22545 |
432 |
421 |
354 |
745 |
720 |
619 |
Table 1 A comparison of the ability of three search engines Mascot (MAS), Sequest (SEQ), and ProteinPilot (PP) to identify deglycosylated proteins in three different urine samples (U1, U2 and U3)
All peptides and proteins were identified at 1% false discovery rate at the peptide level with a minimum of two unique peptides per protein (See Experimental Section). A deglycosite was defined by two criteria: 1) comprising the common N-glycosylation consensus motif (Asn-X-Ser/Thr, where X was any amino acid except proline), and 2) the specific asparagines residue within that motif was identified as the O18-incorporated deamidation derivative (O18Asp). Deglycosylated proteins in this study were defined as the proteins containing at least one identified deglycosite.
N-glycosylation consensus motifs analysis
All N-glycosylation consensus motifs can be classified into three distinct chemical forms: unmodified (Asn-X-Ser/Thr), deamidated (16O-Asp-X-Ser/Thr), and deglycosylated (18O-Asp-X-Ser/Thr). If a specific motif was identified in the deglycosylated form and also identified in either of the other two forms, this indicated that that motif was partially glycosylated.8 When all motifs were compared, evident differences were observed among the results from three search engines (Figure 1a–1c). Similar to its ability to identify more unique peptides, ProteinPilot also detected the largest number of unmodified (Figure 1a) and deamidated motifs (Figure 1b), but surprisingly ProteinPilot identified the smallest number of deglycosylated motifs (Figure 1c).
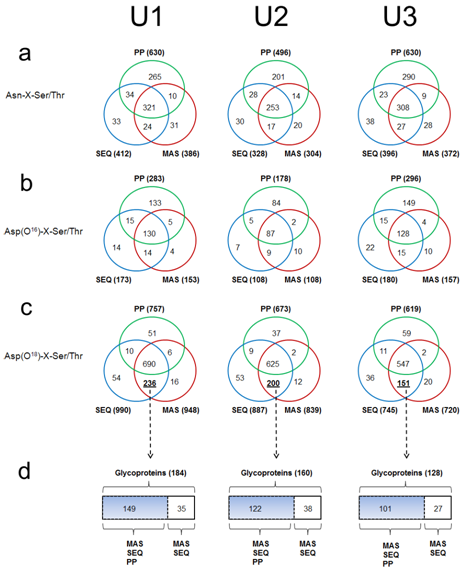
Figure 1 An analysis of the identified N-glycosylation consensus motifs Asn-X-Ser/Thr (where X is any amino acid except proline) from three urine samples by each search engine: Mascot (MAS), Sequest (SEQ), and ProteinPilot (PP).
Figures a, b, c are Venn-Diagram comparisons of unmodified motifs Asn-X-Ser/Thr (a), deaminated motifs Asp(16O)-X-Ser/Thr (b), and deglycosylated motifs Asp(18O)-X-Ser/Thr (c).
Figure 1d is the comparison of a subgroup of glycoproteins which were defined as an exclusive set of deglycosylated motifs identified by both MAS and SEQ and retro-analyzed for their respective parent glycoprotein.
For instance, ProteinPilot identified 630 unmodified and 283 deamidated motifs in sample U1, which was more than 40% higher than those identified by either Sequest (unmodified: 412 and deamidated: 173) or Mascot (unmodified: 386 and deamidated: 153) (Figure 1a). However, ProteinPilot only identified 757 deglycosylated motifs which were far fewer than those identified by Sequest (990) or Mascot (948) (Figure 1c). A large number of deglycosylated motifs were exclusively identified by both Sequest and Mascot but not by ProteinPilot in all three urinary samples (Figure 1c). The consistency of this pattern across all three urine samples indicated that ProteinPilot had a systematically lower sensitivity than the other two engines to detect 18O-incorporated deglycosites.
We propose two possible explanations for this disparity. The first is that the glycoproteins containing these motifs are also identified by ProteinPilot, but the deglycosylated motifs are missed. The alternative is that the glycoproteins containing these deglycosylated motifs have not been identified by ProteinPilot at all. In an attempt to differentiate these scenarios, the set of deglycosylated motifs exclusively identified by both Sequest and Mascot (Figure 1c) were retro-analyzed for their respective parent glycoproteins (Figure 1d). As clearly shown in Figure 1d, the majority of this subgroup of glycoproteins was also identified by ProteinPilot, indicating that these glycoproteins were largely detected by ProteinPilot. Therefore, ProteinPilot’s lower sensitivity in identifying particular deglycosylated motifs appeared to be due to the unique algorithm employed by ProteinPilot. There are myriad underlying reasons why search algorithms disagree on results, including differences in sensitivity, specificity, and fundamental approach to peptide identification. Considering the high level of sophistication and validation each of these engines have received over many years of use, the difference in sensitivity for 18O-incorporated deglycosites is compelling. Notably, ProteinPilot has very few user parameters to optimize, and it was beyond our capability to identify the underlying mechanism of ProteinPilot causing this less sensitive performance, but these Data highlight the need for further research. We would anticipate that future upgrades and optimization of ProteinPilot may resolve this issue. As for Sequest and Mascot, both of them had a similar performance for all three forms of analyzed motifs. Although Sequest seemed slightly better compared to Mascot, the overall differences were insignificant compared to their differences with ProteinPilot.
In this report, three commercial search engines, Sequest, Mascot, and ProteinPilot, were compared side-by-side for their sensitivity in identifying 18O-incorporated deglycosites. When the same search parameters, FDR limits and proteomic identification settings were employed, Sequest and Mascot showed better sensitivity in detecting those deglycosylated sites. ProteinPilot was more likely to identify the consensus motifs as unmodified (Asn) or chemically deamidated (16O-Asp). It is hoped that our results may guide future N-glycoproteomic studies that involve PNGase F catalyzed de-N-glycosylation in an H218O environment.
We would like to thank the Department of Urology at Boston Children’s Hospital for their continued support. The National Institutes of Health grant DK096238 and the Nanji Myelodysplasia Research Fund supported this work. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The author declares no conflict of interest.

©2017 Zhou, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.