MOJ
eISSN: 2374-6920


Research Article Volume 4 Issue 5
Centre for Systems Biology and Bioinformatics, Panjab University, India
Correspondence: Ashok Kumar, Centre for Systems Biology and Bioinformatics, Block-III, Sector-25, Panjab University, Chandigarh-160014, India
Received: August 24, 2016 | Published: December 8, 2016
Citation: Jain M, Gupta K, Kumar A. Text mining and network analysis for discovery of novel genes associated with staphylococcus biofilms. MOJ Proteomics Bioinform. 2016;4(5):311-316. DOI: 10.15406/mojpb.2016.04.00135
Text mining is the process of extracting relevant information from the unstructured data obtained from the published corpora in biomedical literature. A vast amount of literature is available in context of biofilms in PubMed database. It is a very daunting task to extract useful information in the form of genes and proteins from text manually; therefore, text mining approaches are used for this purpose. The extracted relevant information can be appropriately visualized using network biology approaches. Therefore, here we present a framework that would help in discovering novel biomarkers (genes) and gene-drug associations, involved in Staphylococcus biofilm using the available published corpora. We initially utilized COREMINE tool to select biological processes (i.e. biofilm formation, growth, quorum sensing and pathogenesis) and extract genes from PubMed literature database for biofilms. We selected the co-occurring genes for four biofilm processes through network biology approach and validated them by using GO enrichment analysis. AgrB gene was validated as the gene involved in two biological processes i.e. biofilm quorum sensing and pathogenesis. Therefore, structure of AgrB encoded protein (AgrB) was predicted using bioinformatics tools and analysis of ligand binding was done using molecular docking which was further used in blocking quorum sensing and pathogenesis processes. The validation of results obtained from text mining suggests that this approach can be extended to reveal interesting trends and associations among different biological entities related to biofilms and other diseases.
Keywords: text mining, biofilms, network analysis, GO enrichment analysis, AgrB, biomedicine
Advances in technologies related to biomedicine lead to the tremendous data generation through respective researches and discoveries. A wealth of knowledge obtained through these researches is available in the form of published texts. But it is a challenging task to extract relevant information like genes, proteins, diseases, drugs, biological mechanisms etc. from the available unstructured data.1 Integration and analysis of the information obtained from the literature can lead to the new discoveries in medical health, medicine, therapeutic and diagnostic processes.2 Therefore, this information could be easily extracted from the publications by using text mining approaches3 and could be visualized using network biology approaches. Text mining is the branch of data mining which is used to find hidden relevant information from the available published corpora.4 The purpose of text mining here is to process the unstructured textual information related to biofilms in order to extract knowledge like set of biofilm genes, their interactions with biofilm biological processes and associated drug relationship.5 The application of text mining has earlier been seen on the literature of biomedical sciences like respiratory disease, breast cancer, heart diseases etc.6
In the present study we have used the text mining application on the PubMed literature available for Staphylococcusbiofilms. Biofilms are the basically the assembly of various living and reproducing microorganisms that adhere to the surface via glue-like slime and extracellular polymeric substances (EPS).6-8 These microorganisms are various species of bacteria, fungi, algae, yeast, protozoans and other microorganisms.9 Staphylococci are recognized as the most frequent causes of biofilm-associated infections.10 This exceptional status among biofilm-associated pathogens is due to the fact that staphylococci are frequent commensal bacteria on the human skin and mucous surfaces (and those of many other mammals).11 Thus, staphylococci are among the most likely germs to infect any medical device that penetrates those surfaces, such as when being inserted during surgery.12 Therefore, there is need to explore the genes and proteins of Staphylococcus involving in biofilm formation and other biological processes and their related pathways, drugs and other processes associated with these genes. The networks help in appropriate visualization of genetic disorders and all their known gene associations,13 or of drugs and all their known protein targets,14 enabling worthwhile insights into disease and disease therapy.15
Data retrieval
For text mining purpose, the availability of text on the relevant topic is a fundamental necessity.16 This text is made available in the form of published corpora in biomedical literature of biofilms obtained through major resource i.e. PubMed database.17 MeSH major topic “Biofilms” and MeSH term “Staphylococcus” was searched from MeSH database.18,19 Then the literature containing both terms and published during last 10years was obtained.
Exploring Connections from Biomedical Literature
Many tools are available for text mining the biomedical literature. Here COREMINE tool19 was used which is a biomedical text mining tool and extracts all the information like list of genes and proteins, related MeSH terms, processes, diseases, drugs etc. from the literature. It gives the results in an interactive graphical interface. The corpora for biofilms were used to search for genes. The results were downloaded in the form of excel sheet. The results are sorted on the basis of significance score i.e. P-value. This significance score is calculated on the basis of the occurrence of two terms in the same title/abstract of the article. Higher the number of articles lower will be the P-value and thus more significant the connection.20 Four processes i.e. biofilm formation, growth, quorum sensing and pathogenesis was also selected using the same tool.
Co-occurrence of genes
Co-occurrence is a term used in semantics21 that means the occurrence of two terms in a text. If two terms are occurring in same article for the same topic they are more likely to be related to each other. Here we found the co-occurring genes by using network analysis approach. Co-occurrence networks are generally used to provide a graphic visualization of potential relationships. The generation and visualization of co-occurrence networks has become practical with the advent of electronically stored text amenable to text mining.
Network building
All the processes along with their corresponding genes were shown in a network using Cytoscape version 3.3.3.22 Network analysis was done to differentiate the genes on the basis of their significance score. In the network nodes represent the genes and edges represent the significance score. Also all the networks were merged into one and the genes that were co-occurring for multiple processes were selected. Cystoscope is an open source bioinformatics software platform for visualizing molecular interaction networks. Additional features are available as plugins in cystoscope here merged network plug-in was used to merge networks.
GO Enrichment analysis
The genes that were co-occurring for all of the four processes were selected for further enrichment by using GO enrichment analysis.23 This step provides the validation to the genes for their classification to particular processes. These validated genes were further shown in a network corresponding to their validated processes. Again using network analysis genes common for more than one process were found.
Molecular modeling and docking
In this step various bioinformatics tools were used for the structure prediction of selected protein. FALCON@ home tool24 was used for structure prediction. In the Critical Assessment of protein Structure Prediction (CASP11) in the year of 2014, the FALCON@ home based prediction was ranked the 12th in the template-based modeling.17 The ligands that correspond to the selected proteins were found using text mining tool like Coremine and curated database like Stitch4.0. The ligands common from both searches were taken and their binding with corresponding genes/proteins was further validated using molecular docking. Fire Dock (Fast Interaction Refinement in molecular Docking) tool25 was used to refine the results obtained from Patch Dock.26
Data retrieval
There were total 13907 and 87198 PubMed articles published for “Biofilms [MeSH major topic]” and “Staphylococcus [MeSH terms]” respectively and a total of 1568 articles were published in PubMed for both the terms in the previous 10years. These terms were co-occurring in the abstracts and full text of the published articles (Figure 1).
Exploring connections from biomedical literature
Biofilm term was searched and Staphylococcus genes were selected after using text minig approach (Figure 2). There were 25062 connections available for this term in Coremine. Only biological processes and Gene/protein option was selected as shown in Table 1.
Process |
Number of genes |
Range of significance score |
Highly cited gene |
Number of articles |
Less cited gene |
Number of articles |
Formation (P1) |
72 |
1.31E-05-0.03047125 |
ica A |
31 |
isdA |
1 |
Growth (P2) |
82 |
1.14E-04-0.15396536 |
ica A |
30 |
norA |
1 |
Pathogenesis (P3) |
63 |
1.21E-04-0.04513 |
ica A |
14 |
arlR |
2 |
Quorum Sensing (P4) |
21 |
1.58E-04-0.02311349 |
ica A |
10 |
kataA |
2 |
Table 1 Total number of genes for each process and their range of significance scores
Four Biofilm processes were selected in Coremine i.e. Biofilm formation, Growth, Pathogenesis and Quorum sensing. There were a total of only 238 genes corresponding to these four processes. Each connection was associated with a significance score. Thus genes are sorted on the basis of significance score, which is P-value, where low value represents higher significance and thus higher rank as shown in Figure 2.
Network building and co-occurrence of genes
The nodes in the network represent the genes/protein while the source node represents the particular process. The edges are on the basis of significance attribute and highly significant edges are represented in large size. The color of nodes is on the basis of their degree. Lower the degree darker and smaller will be the node. Cystoscope uses statistical measures to make the network more significant. Since there was no overrepresentation of genes in the merged network as shown in Figure 3 therefore there were total 87 unique genes in the dataset. The overrepresentation of genes is due to their involvement in more than one process.
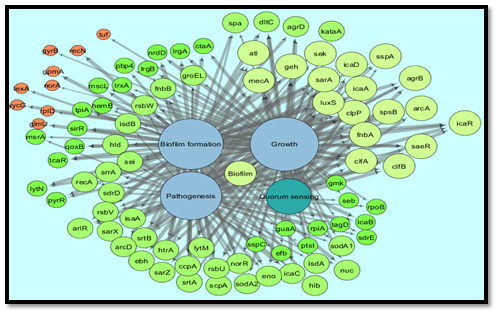
As this dataset was obtained using text mining so these overrepresented genes were actually the genes that were co-occurring in the titles/abstracts of the articles for more than one process. In this way the co-occurring genes can be selected from this network with degree layout which showed that the frequency of nodes sharing two neighbors is higher than those sharing four neighbors. It means that there were nodes representing genes that were involved in all four biological processes. Those genes involved in all of the four processes were selected using network analysis and degree layout. There were total 18 genes, obtained from the above merged network, that appear in all of the four processes of biofilm Figure 4.
GO enrichment analysis
The selected genes were validated by using GO enrichment analysis. List of 18 genes were pasted under Enrichment analysis input box. Biological process was selected as it reduces the analysis time by selecting one process at a time. The pie chart shown in Figure 5 is prepared using the data obtained from GO enrichment analysis.
Out of 18 genes 16 genes were taken by GO consortium. Out of these 16 genes 2 were unclassified and rest classification of genes are shown in the form of pie chart. This enrichment validates that out of 18 genes, as obtained from text mining, 7 genes were there in Pathogenesis process while 2 genes were validated to be involved in quorum sensing process. Out of these 9 genes we were interested to find further common genes that could target both the processes simultaneously.
From this network Figure 6 we found AgrB gene that was involved in both processes.27 This could be an interesting gene because by targeting this gene’s activity both processes can be inhibited simultaneously.
Structure prediction and molecular docking
After finding the potential gene the next step was to predict the structure of gene encoded protein and perform molecular docking with common ligands obtained from both text mining and curated database searches. The structure of AgrB gene encoded protein i.e. AgrB (Accessory gene regulator protein B) was predicted using the online tool FALCON@ home. The Fasta sequence of protein with Uniprot id P0C1P7 was pasted in the FALCON@ home. It performs the template based modeling. The structure of AgrB protein was predicted using template with PDBid 1R8S. The alignment file of 1R8S and AgrB was provided by FALCON and the alignment score is 23.9459. The ligands to be docked was again extracted using both approaches i.e. text mining and database search. These searches showed 3-oxo-C12-HSL was highly significant using database search and Homoserine Lactone was highly cited drug as shown by text mining Figure 7.
The structures of both the ligands were obtained from Drugbank. Also on comparing the properties of both ligands it was found that L-Histidine had more hydrophobic character than Homoserine Lactone (HSL) as logP value of former is highly negative. Moreover, number of Hydrogen bond donor and acceptor were also more in case of L-Histidine which means it binds with receptor more efficiently than HSL. So docking was performed using Patch Dock and further refined the results of docking using Fire Dock. Here three docking poses for both HSL and L-Histidine bound with AgrB protein were compared. The results were compared on the basis of global energy which predicts the stability of the complex. In both results pose1 had the lowest global energy. We also plotted a graph of comparative global energies for both docking poses. It was found that the complex of AgrB and L-Histidine was more stable than with HSL which was otherwise highly cited ligand for AgrB (Figure 8).
The present work is an application to text mining and network analysis approach. In the present work we started with 238 genes obtained using text mining which were then sorted down to 9 on the basis of their enrichment and validation processes. The co-occurring and validated genes found more interested than the rest as it can provide the way to novel receptor prediction that could target multiple processes simultaneously. Only 9 genes were validated for two important biological processes i.e. Quorum Sensing and Pathogenesis. These two processes play an important role in biofilm development and survival. Thus genes responsible for these processes are also of great importance. After further analysis, AgrB gene was found to be involved in both processes and blocking of this gene encoded protein might suggest the inhibition of both processes simultaneously. Again we found drugs for AgrB protein using text mining and found a less cited ligand L-Histidine that bound more efficiently to AgrB protein than HSL, a highly cited ligand for AgrB. Molecular docking and other curated databases were also used for this purpose. Thus, through text mining we could discover those relations that were less or never been discovered before.28
Authors are thankful to Vice Chancellor of Panjab University, Chandigarh and Coordinator of Centre for Systems Biology and Bioinformatics for providing all the facilities to carry out this work. We would also like to acknowledge the support provided by online tools, servers, databases and softwares for successful accomplishment of our work that is
PubMed Database (http://www.ncbi.nlm.nih.gov/pubmed)
Gene Ontology Consortium (http://geneontology.org/)
Coremine (https://www.coremine.com/medical/)
FireDock (http://bioinfo3d.cs.tau.ac.il/Fire Dock/)
Cytoscape (http://www.cytoscape.org/release_notes_3_3_0.html)
PatchDock (http:// bioinfo3d.cs.tau.ac.il/PatchDock/)
DrugBank (http://www.drugbank.ca/) and FALCON@home (http://protein.ict.ac.cn/FALCON/).
The author declares no conflict of interest.

©2016 Jain, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.