MOJ
eISSN: 2374-6920


Research Article Volume 2 Issue 3
Department of Biological Sciences, Charles E. Schmidt College of Science, Florida Atlantic University, USA
Correspondence: Ramaswamy Narayanan, Department of Biological Sciences, Charles E. Schmidt College of Science, Florida Atlantic University, 777 Glades Road, Boca Raton, FL 33431, USA, Tel +15612972247, Fax +15612972247
Received: May 28, 2015 | Published: July 24, 2015
Citation: Narayanan R. Druggable vitiligo genome: a fast track approach to take the genome wide association to the clinic. MOJ Proteomics Bioinform. 2015;2(3):102-110. DOI: 10.15406/mojpb.2015.02.00050
Vitiligo, a skin depigmentation disorder, affects 0.5 to 1% of the population around the globe. While not life threatening, the disorder is associated with serious psychological trauma. Currently no known cure is available and the precise etiology is unknown. Genome-Wide Association Studies (GWAS) may provide clues to better understanding the disorder for the development of novel therapeutics. Mining the GWAS databases resulted in the identification of 51 Vitiligo-associated genes (VAG) encompassing protein-coding sequences, noncoding RNAs and pseudogenes. A druggable class of proteins including enzymes, transporters, transcription factors and secretome products was part of the VAG. The VAG were also genetically linked to autoimmune, cancer, cardiovascular, inflammation, infections and neurological diseases. Two genes, Interleukin 2 receptor alpha (IL2RA) and Tyrosinase (TYR) are FDA approved targets. Unique population-specific genes were identified in the GWAS databases. Chemogenomics approaches identified 246 compounds targeting the VAG. Key pathways involving the VAG’s mechanism included apoptosis, endocrine, immune, infection, metabolic, neuronal and transcription factor signaling. Five lead targets, four enzymes and one transporter, emerged from this study with bioactive drug-like compounds (<100nM). The VAG were linked to diverse environmental factors including antioxidants, DNA damage, oxidative stress and UV. Drug bank compounds encompassing anti-infectives, anti-neoplastics, immune modulators and nutraceuticals/ supplements were identified as targets for the VAG. The FDA approved drugs from the VAG studies can be repurposed and off label use can be developed for the treatment of Vitiligo.
Keywords: chemogenomics, chemoinformatics, druggable genes, human genome, ligand binding, protein 3d structures, response to therapy, skin disorders
canSAR, integrated cancer drug discovery platform; chEMBL, database of bioactive compounds at the european bioinformatics institute of the european molecular biology laboratory (EMBL); FDA, us federal drug administration; GWAS, genome-wide association studies; VAG, vitiligo-associated genes; PDB, protein database; RO5, rule of five
Vitiligo is an acquired depigmenting disorder of the skin and the mucous membrane characterized by loss of melanocytes, exocrine cells from the basal layer of the Epidermis and the matrix portion of the hair bulb.1,2 The disease is seen at a frequency of 1% of the world population.3 Vitiligo is classified into four types based on the distribution of the hypopigmented lesions: non-segmental Vitiligo (NSV), segmental Vitiligo (SV), mixed NSV and SV and unclassified types including focal, multifocal asymmetrical non-segmental and mucosal at one site.2 The NSV subtypes include focal at onset, mucosal, acrofacial, generalized and universal. Generalized Vitiligo occurs later in life at sites sensitive to pressure and is often progressive with flare-ups. SV usually begins in childhood most commonly in the face and remains stable.3 The white skin patches associated with Vitiligo have been recognized over thousands of years.4
Despite the long history, the etiology remains unknown. In the recent years, numerous theories have been put forward.1,5-12 Current evidence encompasses four main theories: the autoimmune hypothesis, the neural hypothesis, the self destruct hypothesis and the growth factor defect hypothesis.13 However, none of these have been conclusively proven to date. It is highly likely that Vitiligo is a polygenic trait and a convergent theory, combining elements of different theories across a spectrum of expression, is the most accurate etiology.7 The association of Vitiligo with other known autoimmune disorders such as Addison’s Disease, Hashimoto’s Thyroiditis, Pernicious Anemia and Alopecia Areata lend credence to the autoimmune theory of disease.14
While non-life threatening, the disfigurement associated with Vitiligo causes serious emotional stress and depression and interferes with the quality of life.15,16 No known cure is available and current treatment options are limited. Diverse therapeutic modalities are in current use, including topical (corticosteroids, vitamin D derivatives, calcinurin inhibitors, prostaglandin E 2, Pseudocatalase), phototherapy [psoralen plus UV-A (PUVA), psorolen with sun light (PUVAsol), surgical techniques, Excimer laser, and combinations of topical therapy with light treatment.1,17-19 Other medicines being tried include alternative medicines such as ginkgo biloba,20,21 topical fluorouracil,22 minioxidil,23 oral L-phenylalanine,24 homeopathy and aurvedic medicine17,25 and depigmentation with monobenzyl ether.26 The success rate with all of these drugs is variable and uncertain. At the present time the only FDA approved treatment is the use of Excimer laser (308nm).27,28 Currently, even if the treatment is efficacious, often the depigmentation recurs. A maintenance therapy is often required.19 In view of the large number of people affected around the globe by Vitiligo, novel therapeutic approaches as well biomarkers for identifying at risk individuals are urgently needed.
Currently 53 clinical trials are underway for the treatment of Vitiligo (clinicaltrials.gov). The International Clinical Trial Registry (WHO) platform records 28 clinical trial records for Vitiligo. The European Union Clinical Trials Register shows 11 clinical trial records currently underway in Europe. These trials largely revolve around light treatment, excimer laser, topical steroids and calcineurin inhibitors, statins, anti oxidants, vitamins, ginkgo biloba supplements, skin grafts, melanocytes and keratinocytes transplantation.
The GWAS dataset for the Vitiligo-associated phenotypes are becoming available.29-34 Reasoning that the phenotypic polymorphic traits associated with the Vitiligo genes might provide an attractive starting point for drug targets discovery, the GWAS databases were mined for VAG. Fifty-one genes were identified from these studies encompassing druggable targets. Chemogenomics approaches identified highly active small molecular weight compounds (<100nM) for five of these targets, which include enzymes and a transporter. FDA approved and experimental drugs as well as nutraceuticals were identified targeting the VAG. Repurposing these approved drugs may offer a therapeutic benefit to the Vitiligo patients.
The bioinformatics and proteomics tools used in the study have been described.35-37 The GWAS catalogue38 and the NCBI Phenome-Genome Integrator, PheGenI39 were used to create a working list of VAG. Characterization of the VAG was performed using the GeneALaCart, GeneAnalytics and VarElect data mining tools from the GeneCards.40 The VarElect is a Variant Election application for disease/phenotype-dependent gene variant prioritization. VarElect combines information within the LifeMap Knowledgebase of GeneCards, MalaCards, the human disease database, LifeMap Discovery, the regenerative medicine, and PathCards, the unified human biological pathways database.
In addition, Gene Analytics Powered by LifeMap’s Gene Cards Suite integrated knowledgebase, which utilizes data from >120 select sources was also used to batch analyze the VAG for gene expression-based analysis. Comprehensive background information on the VAG was obtained by using the GeneALaCart tool from the GeneCards for Gene Ontology, super pathways and drug bank compounds.
The protein annotation and chemical structure-based mining was performed using the canSAR 2.0 integrated knowledgebase, a publically available database.41,42 The browse canSAR section was used and the Vitiligo-associated proteins were batch analyzed for protein annotations, 3D structures, compounds and bioactivity details. The protein 3D structure template models-related information was obtained from the Swiss Protein Database.
The chemical structures were obtained from the chEMBL.43 Comprehensive gene annotation for the VAG was established using the GeneCards,40 the DAVID functional annotation tool,44 and the UniProt45 databases. FDA approved drug related information was obtained from the Drug-Bank.46 Protein expression was verified using the Human Protein Map,47 ProteomicsDB48 and the Multi Omics Protein Expression Database.49,50
The canSAR compounds link for genes has diverse filters such as activity and assay types, concentrations, molecular weight, RO5 violations, prediction of oral bioavailabilty and toxicophores. Putative drug hits were filtered from the canSAR datasets for the VAG-associated genes using Lipinski’s rule of five (also known as Pfizer’s rule of five), RO5. The RO5 is a rule of thumb to evaluate druggableness or to determine whether a compound with a certain pharmacological or biological activity possesses properties that would make it a likely orally active drug in humans.51-53 Highest stringency was chosen for the RO5 violation (value=0). Protein 3D structural homology was set at 100%. The ligand-based druggability score was chosen at >60% confidence. Drugs with IC50values (<100nM), inhibitory activities and Ki values were chosen for the canSAR output. Toxicophore negative was chosen to filter the hits for toxicity associated compound structures54
Vitiligo-associated proteins from the GWAS studies
The GWAS databases (the GWAS catalogue at European Molecular Biology Laboratory-European Bioinformatics Institute -EMBL-EBI) and the NCBI PheGenI were mined for the query term Vitiligo. The output from these two databases was merged and duplicate entries were removed. This resulted in the identification of 51 Vitiligo genes including protein coding genes, noncoding RNAs and pseudogenes (Figure 1 & Supplemental Table S1). As a class, enzymes were the largest amongst the VAG (kinase, ligase, helicase, tyrosine phosphatase, granzyme B, caspase 7 and ribonuclease T2). Other druggable classes of proteins included transporters (Phospholipid, amino acid, nucleoside and Choline) and receptors (Mast cell immunoreceptor signal transducer, RIG-I-like receptor 2, Interleukin-2 receptor subunit alpha, Toll-interleukin-1 receptor domain-containing adapter protein and Suppressor of T-cell receptor signaling 2).
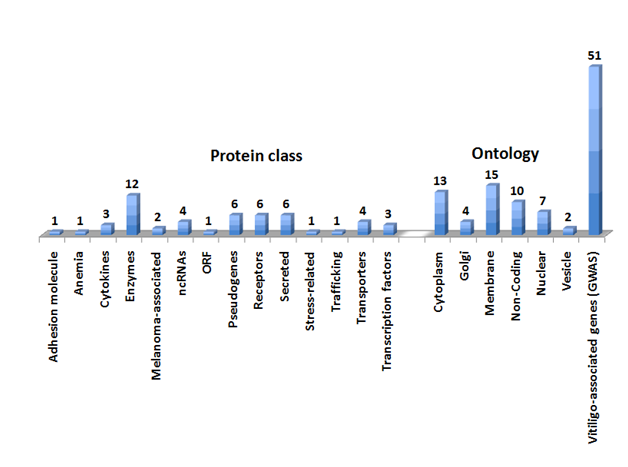
Secreted proteins included Complement C1q tumor necrosis factor-related protein 6, Prolyl endopeptidase, Granzyme B, Ribonuclease T2, calcium-binding protein 2 and Thyroglobulin. Two of the Vitiligo proteins were associated with melanoma (Prolyl endopeptidase FAP and Interferon-induced helicase C domain-containing protein 1). The Gene Ontology analysis revealed distinct subcellular locations of the Vitiligo genes including cytoplasmic, nuclear, golgi, vesicular, secreted and membrane bound proteins (Figure 1 & Supplemental Table S1).
Vitiligo proteins in other diseases and population distribution
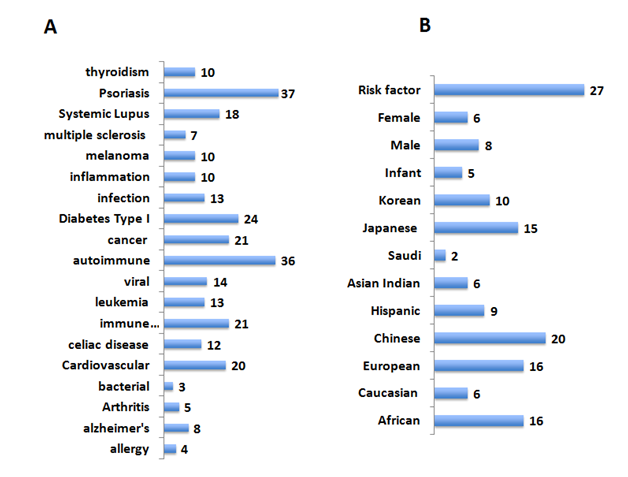
Vitiligo is often seen in patients with autoimmune and inflammatory diseases.3,55 It is, however, unclear whether such an occurrence is a coincidence. In order to identify common pathways and possible links to diverse diseases and Vitiligo, the VAG were analyzed for phenomo-genome genetic association for disease-related variants using the NCBI PheGenI and VarElect datamining tool from the GeneCards (Figure 2, panel A). The VAG association was seen with autoimmune diseases, allergy, cancer, inflammation, infections and neurodegenerative diseases. The availability of the GWAS datasets from around the globe with the 1000 genome project56 enabled the study to expand into population relevance (Figure 2, panel B). Twenty-seven of the VAG were found to be risk factors for diverse diseases including skin diseases (Supplemental Table S1).
Distinct genes related single nucleotide polymorphisms (SNPs) were associated with unique population (Supplemental Table S1). These included patients from Japan (heterogeneous nuclear ribonucleoprotein A1 pseudogene 2|HNRNPA1P2), Africa (solute carrier family 44, member 4|SLC44A4, zinc finger, MIZ-type containing 1|ZMIZ1) and Europe (potassium channel, subfamily K, member 12|KCNK12, mitochondrial ribosomal protein S17 pseudogene 7| MRPS17P7, SPARC related modular calcium binding 2|SMOC2 and toll-like receptor adaptor molecule 1|TICAM1).
Association of some of the VAG is gender-specific: males (ATPase, aminophospholipid transporter, class I, type 8B, member 1, ataxin 2 and RPGRIP1-like) and females (tyrosinase). Generalized Vitiligo and psoriasis shared 37 genes from the VAG list. Interestingly, an enzyme, ADP-ribosylarginine hydrolase, was found to be associated with segmented, but not generalized Vitiligo. Variants related to these unique genes may provide a Vitiligo fingerprint or biomarkers potential to monitor Vitiligo in a population specific manner.
Vitiligo genes: types of mutations
The Vitiligo genes were investigated for the types of mutations occurring in all tissues versus skin tissue (Figure 3). In the skin tissue germ line, pathogenic and causative mutations were seen for three genes 1) Fanconi anemia, complementation group A, 2) solute carrier family 29 (equilibrative nucleoside transporter), member 3 and 3) tyrosinase.
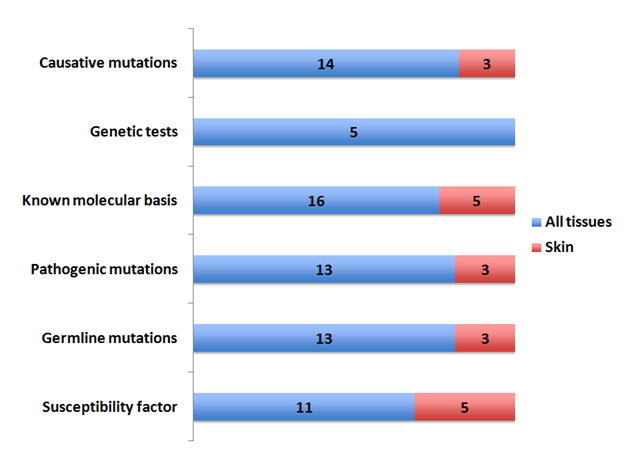
Five genes predictive of susceptibility to skin diseases/risk factors were also identified in these studies. These included granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1), major histocompatibility complex, class 1B, interferon induced with helicase C domain 1, protein tyrosine phosphatase, non-receptor type 22 (lymphoid) and fibroblast activation protein, alpha.
Pathways mapping of the Vitiligo genes
A Meta analysis tool, Gene Analytics, from the GeneCards was used to establish pathways implicated with the Vitiligo genes (Table 1). Consistent with the complex multifactorial etiology of Vitiligo,7 the VAG were found to be associated with pathways involving immune function, infectious diseases, endocrine function, apoptosis and survival, transcription factor signaling, metabolic and neural pathways. A group of genes common to these diverse pathways encompasses caspase 7, apoptosis-related cysteine peptidase, granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1), interleukin 2receptor, alpha and tyrosinase. Vitiligo is often associated with diverse environmental stress (e.g. DNA damage, stress, UV exposure). However, the precise role of any of these in the etiology of Vitiligo is unclear. Using the GeneAnalytics Meta Analysis tool, the variants of the VAG were analyzed for association with diverse factors (Supplemental Table S1). Distinct patterns of polymorphic variants were found to be associated with cell death (apoptosis, differentiation), steroids, stress-related, DNA damage (reactive oxygen, UV, phototherapy), melanocytes and vitamins and supplements.
Super Pathways (Gene Anaytics) |
Vitiligo-Associated Genes (VAG) |
Immune |
|
Lymphocyte Signaling |
BACH2, GZMB, IFIH1, IL2RA, PTPN22 |
Allograft Rejection |
CASP7, GZMB, HLA-B, IL2RA, TG |
IL-15 Signaling and Its Primary Biological Effects in Different Immune Cell Types |
GZMB, IL2RA |
IL12-mediated Signaling Events |
GZMB, IL2RA |
Downstream Signaling in Naive CD8+ T Cells |
GZMB, IL2RA |
Infection |
|
Influenza A |
HLA-B, IFIH1, IL2RA, TICAM1 |
Pertussis |
CASP7, TICAM1 |
Endocrine |
|
Thyroxine (Thyroid Hormone) Production |
TG |
Apoptosis |
|
Activation of Caspases Through Apoptosome-mediated Cleavage |
CASP7 |
Apoptosis and Survival Caspase Cascade |
CASP7, GZMB |
Apoptosis (WikiPathways) |
CASP7, GZMB |
Activation of BH3-only Proteins |
CASP7, GZMB |
Granzyme Pathway |
CASP7, GZMB |
Signaling |
|
NF-kappaB Signaling |
IFIH1, TICAM1 |
S-1P Stimulated Signaling |
CASP7, CDH23 |
Metabolic |
|
Riboflavin Metabolism |
TYR |
(S)-reticuline Biosynthesis II |
TYR |
Neuronal |
|
Dopamine Metabolism |
TYR |
Neurotransmitter Uptake and Metabolism In Glial Cells |
SLC1A2 |
Parkinsons Disease Pathway |
ATXN2, CASP7 |
Table 1 Pathway mapping of the vitiligo genes. Super Pathways involved with the VAG analyzed by the GeneAnalytics Meta Analysis tool (GeneCards) are shown
Vitiligo genes in the secretome
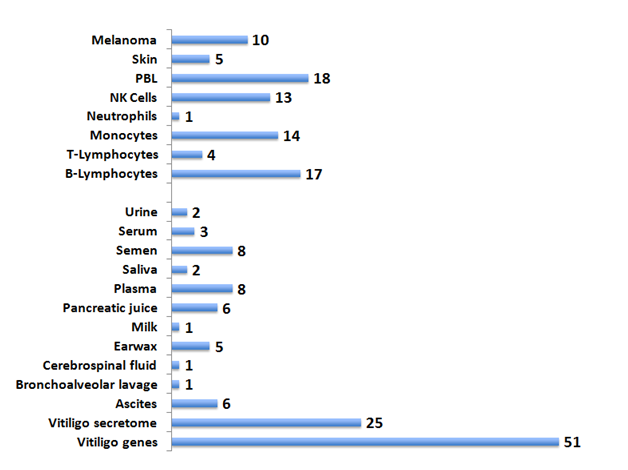
The availability of the human proteome data sets47,48,57 allows the analysis of the disease-related genes in diverse body fluids. The human secretome encompasses proteins secreted by both the classical (signal peptide cleavage) and non-classical secretory pathways such as shed receptors and enzymes.58–61 Identifying the Vitiligo proteins secreted in the body fluids is crucial to developing a biomarker potential for the genes. Hence the proteomics database, the MOPED50 was batch analyzed for the 51 VAG (Figure 4 & Supplemental Table S1). The VAG protein expression was detected in diverse body fluids including ascites, blood, cerebrospinal fluid, earwax, milk, pancreatic juice, saliva, semen and urine. The proteins detected in fluids such as 1) earwax (BTB and CNC homology 1, basic leucine zipper transcription factor 2|BACH2;DEAD (Asp-Glu-Ala-Asp) box helicase 6 |DDX6; Fanconi anemia, complementation group A |FANCA; ribonuclease T2|RNASET2 and solute carrier family 44, member 4 |SLC44A4), 2) milk (HLA-B), saliva (major histocompatibility complex, class I, B|HLA-B and ribonuclease T2| RNASET2) and 3) urine (major histocompatibility complex, class I, B |HLA-B; arginine-glutamic acid dipeptide (RE) repeats |RERE) could provide a basis for non-invasive biomarker for prediction of risk as well as response to therapy.
Chemogenomics of Vitiligo-associated genes
In order to develop a drug therapy potential for the Vitiligo proteins, the 51 genes were analyzed using the CanSar protein annotation tool (Supplemental Table S1). Druggability of these genes was inferred from the 3D structures and the ligand binding scores (>70% confidence). Thirty-seven Vitiligo proteins were predicted to be druggable by the canSar annotation tool (Figure 5 & Supplemental Table S1). Among these hits, 18/37 proteins had a significant ligand binding druggability percentile rank (>70%). These hit targets encompassed enzymes, transporters, receptors and secretome. Bioactive small molecular weight (<500) compounds (n=363, <1uM) were identified targeting the Vitiligo proteins. A significant number of these drug-like compounds were predicted to be non- toxic and orally bioavailable (no toxicophore, RO5 value - zero). Highly active compounds (n=46, <10nM) for three unique targets (Granzyme B|GZMB; HLA class II histocompatibility antigen|HLA-B and Tyrosine-protein phosphatase non-receptor type 22 |PTPN22) were identified.
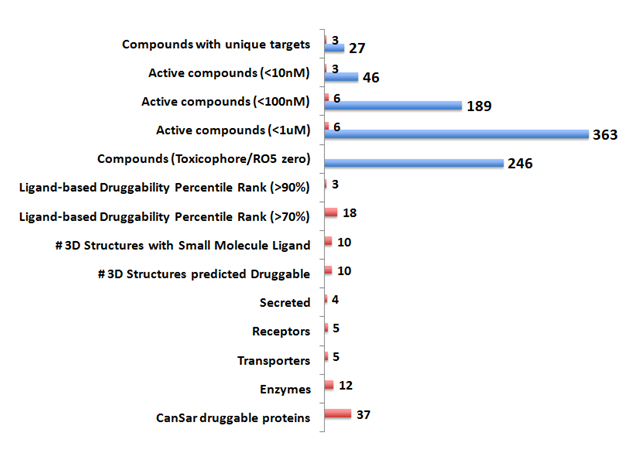
Five lead targets emerged with drug-like compounds (<100nM) bioactive in inhibitory assays (Supplemental Table 2). These leads included four enzymes (caspase 7-CASP7; fibroblast activation protein, alpha-FAP; granzyme B- GZMB and tyrosinase- TYR) and one transporter protein (solute carrier family 1 (glial high affinity glutamate transporter), member 2- SLC1A2). The five chEMBL compounds identified showed a high degree of selectivity to the target proteins. Two of these leads (chEMBL303944 and chEMBL24152) targeted unique enzymes, granzyme B |GZMB and Tyrosinase|TYR respectively.
Drug bank compounds for the Vitiligo genes
Repurposing existing FDA approved drugs for other indications is becoming increasingly attractive.62–64 A powerful example of this concept was recently demonstrated for the Ebola virus disease therapeutics.36,65 Association of genes with multiple diseases opens up avenues for expanding the repurposing strategy. Hence, the 51 Vitiligo genes were screened against the drug bank of compounds using the GeneALaCart Meta analysis tool of the GeneCards. The output was also compared against the FDA database for approved and experimental drugs, nutraceuticals and supplements (Phase I to Phase III). Additional hints of drug therapy use for the Vitiligo genes were obtained from the recently described proteomics reports encompassing putative drug therapy potential for the human proteome.57,66 Two genes, Interleukin-2 receptor subunit alpha (immune suppression, transplant rejection and cancer) and tyrosinase (dermatology, acne, antimicrobial, melanocytes depigmentation, DNA synthesis inhibition), are FDA-approved drug targets (Table 2).
Vitiliogo Associated Genes (VAG) |
Drugs (Drug Bank Compounds) |
Indications |
|
|
|
ADPRH, CASP7, GZMB, HLA-B, IL2RA, SLC1A2, TG, TYR |
Aspartate |
Nutraceutical |
ADPRH, CASP7, FANCA, HLA-B, IL2RA, SLC1A2, TG, TYR |
H2o2 |
Infection|anticancer |
ADPRH, GZMB, HLA-B, IL2RA, PTPN22, SLC1A2, TG, TYR |
Arginine |
Nutraceutical |immune responsesl tissue repair |
ADPRH, CASP7, GZMB, IL2RA, PTPN22, SLC1A2, TG, TYR |
Glutamate |
nutritional supplementation |
ADPRH, CASP7, GZMB, HLA-B, SLC1A2, TG |
Lactate |
dry, scaly skin (xerosis) and ichthyosis vulgaris and for temporary relief of itching associated with these conditions |
GZMB, TYR |
Concanamycin A |
osteoporosis|infections|anticancer |
ADPRH, HLA-B, IL2RA, TG, TYR |
Polysaccharide |
Nutraceutical| treatment and prevention of osteoarthritis, by itself or in combination with chondroitin sulfate |
ADPRH, TG, TYR |
Glucosamine |
|
|
||
|
||
osteoarthritis|dietary supplement |
||
GZMB, IL2RA, TG |
IVIG |
immunodeficiencies, as well as autoimmune and inflammatory disorders. |
HLA-B, IL2RA |
Phycoerythrin |
Anticancer |
HLA-B, TYR |
Castanospermine |
Infection |
IL2RA |
basiliximab| daclizumab, Zenapax|Denileukin diftitox|aldesleukin |
Immunosuppression| For prophylactic treatment of kidney transplant rejection| treatment of cutaneous T-cell lymphoma| treatment of adults with metastatic renal cell carcinoma. |
IFIH1 |
Hyaluronan |
osteoarthritis|plastic surgery|ophthalmology|interstitial cystitis |
ADPRH, HLA-B, IL2RA, SLC1A2, TG, TYR |
Adenylate |
nutritional supplementation| dietary shortage or imbalance |
ADPRH, IL2RA |
Hydroxychloroquine |
|
|
||
|
||
|
||
malaria|discoid and systemic lupus erythematosus, and rheumatoid arthritis. |
||
GZMB, HLA-B, IL2RA, TG, TYR |
Paraffin |
emulsifying agent for cosmetic creams and lotions, mineral oil and paraffin wax emulsions, as a biological buffer, and used as an alkalizer. |
CASP7, GZMB, HLA-B, IL2RA, TG |
Cyclosporin A |
|
treatment of transplant (kidney, liver, and heart) rejection, rheumatoid arthritis, severe psoriasis. |
||
ADPRH, HLA-B |
Busulfan |
|
|
||
chronic myelogenous (myeloid, myelocytic, granulocytic) leukemia|immunosuppressive effect on bone marrow. |
||
ADPRH, CASP7, TG |
Adp ribose |
|
ADPRH, TYR |
Chitosan |
cosmetic |
GZMB, HLA-B, IL2RA, TICAM1 |
Rantes |
acute and chronic inflammation |
ADPRH, ATXN2, RERE, SLC1A2, TG |
Glutamine |
Nutraceutical |
GZMB, TG |
Chromium |
Nutraceutical |
CASP7, FANCA, IL2RA, TG, TYR |
Cisplatin|5FU|Methotrexate |
|
|
||
Anticancer |
||
SLC44A4 |
Choline |
Nutraceutical| CNS disorders |
TYR |
Azelaic Acid|Monobenzone|Mimosine|NADH |
Topical|Dermatology|Neutraceutical |
Table 2 Repurposing approved drugs for vitiligo therapy: Drug bank data
The drug bank compounds encompassing FDA approved drugs and Nutraceuticals indications for the VAG are shown. Skin indications are bolded. FDA drug targets are underlined.
Abbreviations: FDA: Federal Drug Administration; IVIG: Intravenous immunoglobulin; RANTES: regulated on activation, normal T cell expressed and secreted- CCL5
In addition, other approved drugs implicated with the Vitiligo associated genes included 1) anti-neoplastics (Hydrogen peroxide, Concanomycin A, Phycoerythrin, basiliximab, daclizumab, Zenapax, Denileukin diftitox, aldesleukin, Busulfan, Cisplatin|5FU|Methotrexate), 2) antiinfectives (Hydrogen peroxide, Concanamycin A, Castanospermine, Hydroxychloroquine), 3) anti-inflammatory (Intravenous immunoglobulin, Rantes), rheumatoid arthritis (Hydroxychloroquine, Cyclosporin A) and 4) Nutraceticals and supplements (Aspartate, Arginine, Glutamate, Glucosamine, Adenylate, Chromium, Choline, Polysaccharide, chondroitin sulfate). Of particular interest was the discovery of Vitiligo genes associated with dermatological applications including (ADP-ribosylarginine hydrolase |ADPRH; caspase 7 |CASP7; granzyme B |GZMB; major histocompatibility complex, class I, B| HLA-B; solute carrier family 1 (glial high affinity glutamate transporter), member 2 |SLC1A2; thyroglobulin |TG and tyrosinase| TYR; interferon induced with helicase C domain 1 |IFIH1 and interleukin 2 receptor, alpha |IL2RA).
Vitiligo, a complex acquired skin disorder, has been known for centuries.4 Various theories including autoimmune, inflammation, melanocytes, neuronal, stress and environmental factors have been put forward over the last decade.3,6,7,9 The cause and etiology, however, remain unclear. It is likely that a combination of factors is responsible in susceptible individuals. The disease, while not life threatening, affects a large number of people around the world (1%) across racial divide, gender and ages.1,17 The psychological trauma associated with Vitiligo exerts a considerable toll on the patients in view of visible skin abnormality particularly in exposed areas such as the face.16
Currently a cure remains elusive and treatment options are very limited and their effects often only temporary. In fact, the only FDA approved treatment is the use of UV, 308nm Excimer Laser.28 Other treatment options include use of topical calcineurin inhibitors, steroids and vitamins, phototherapy with UV-B, oral antioxidants, herbal supplements and nutraceuticals, depigmentation and camouflaging using cosmetics.3,5,17,35 These treatments have varying degrees of success and the disease remains unconquered. Encouragingly, a number of clinical trials are underway (Supplemental Table S1). Discovery of novel targets for druggableness and off label use of existing drugs is an attractive option to tackle this disease. Experience is emerging for repurposing existing, FDA approved drugs for other diseases.36,62,67 Since these drugs are already in the clinic and their level of toxicity is known, it is relatively easy to develop off the label use for at least some of the drugs, if a molecular link is demonstrable.
In recent years, a huge amount of genetic information has been generated using the GWAS approach.68–73 With the recent advances in human protein expression datasets47,49,57 protein 3D structure availability for the majority of human proteins, and with the use of powerful chemogenomics tools to predict druggableness,42,74–76 accelerated drug discovery starting from the genome is becoming a reality.
Reasoning that a working database of genes associated with Vitiligo can be an attractive starting point for novel diagnostics and therapeutics, the GWAS datasets were mined for VAG (Supplemental Table S1). Fifty-one genes showing polymorphic genetic association with Vitiligo encompassing protein coding and non-coding sequences were identified in this study. Encouraged by the preliminary results that the VAG included putative druggable targets including enzymes, transporters, transcription factors and receptors,58 a comprehensive analysis was undertaken.
Two of the 51 genes (Tyrosinase, TYR and Interleukin-2 receptor subunit alpha, IL2RA) are on the FDA approved target list.46,57 The IL2RA targeted drugs are immune modulators, which inhibit cytokines production and reduce pro inflammatory cytokines; this imples a strong possibility of therapeutic advantage for Vitiligo. The second FDA approved target, TYR targeted drugs, includes anti neoplastics, antibiotics, anti-inflammatory, antioxidants, neurotrophic, neutraceuticals and vitamins. The strong Vitiligo association seen with these two approved targets and the plethora of drugs and nutraceuticals available against these targets provide a strong rationale for testing for off label use.
The genes associated with Vitiligo were also found to be associated with various autoimmune diseases by the NCBI PheGenI tool (Systemic lupus, multiple scelerosis, diabetes Type I, hypothyroidism, rheumatoid arthritis) as well as with cancer, inflammation, infections and neurological disorders. These results are consistent with the prevailing theories that imply diverse etiology.1,3,6,7,17,55 In addition, pathway mapping from the Gene Analytics meta analysis tool also provided corroborating evidence for the involvement of immune, infection, endocrine, apoptosis, NF-kB transcription factor signaling, metabolic perturbation and neuronal pathways. Specific as well as common genes from the VAG are implicated with these pathways. Drugs implicated in these pathways may provide valuable additions to the therapy of Vitiligo. In particular, drugs targeting NF-KB transcription factor, diverse apoptotic caspase-related cascade inhibitors, cytokine signaling inhibitors and inhibitors of neuronal pathways including Dopamine, Parkinson’s and neurotransmitter uptake inhibitors, may be of therapeutic benefit to the Vitiligo patients.
While diagnosis of Vitiligo is relatively easy due to the visible skin abnormalities (white patches), prediction of risk of development of the disease, in particular at a early age as well as monitoring a response to therapy urgently need genetic markers that can be easily monitored. To this end, secreted proteins in body fluids (the secretome) offer a biomarker potential. Half of the VAG proteins are detected in the body fluids (25/51). Five of these secreted proteins were also detected in the skin tissue (ADP-ribosylarginine hydrolase |ADPRH, DEAD (Asp-Glu-Ala-Asp) box helicase 6| DDX6, LIM domain containing preferred translocation partner in lipoma| LPP, ribonuclease T2| RNASET2 and thyroglobulin |TG). In particular, the proteins detected in the body fluids such as ear wax, saliva, semen, milk and urine offer a means to develop noninvasive biomarkers; the proteins detected in the blood and serum can provide a less invasive detection capability. If a definitive link with Vitiligo is established for these biomarkers, they could provide a response to therapy potential for dermatologists to follow. Protein-based nanochips can be easily developed to simultaneously monitor these biomarkers.77
The current GWAS data allowed for segregation of VAG linked polymorphic variants across diverse human DNAs (Chinese/Korean/Japanese; Asian Indians, Saudi; Hispanics; Africans and Caucasians and Europeans). Unique as well as common VAG markers were identified in the study. The unique markers included protein coding sequences as well as pseudogenes and ncRNAs. These markers deserve additional studies and currently ongoing diverse clinical trials could benefit by understanding the expression profile of these genes. Numerous drug-like compounds were identified in the study using chemogenomics approaches. These compounds were predicted to be nontoxic (lack of toxicophore), orally bioavailable (RO5 value zero) and bioactive at nM concentrations. Five lead targets and bioactive compounds were identified in the study (caspase 7, apoptosis-related cysteine peptidase |CASP7, fibroblast activation protein, alpha| FAP, granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |GZMB, solute carrier family 1 (glial high affinity glutamate transporter), member 2 |SLC1A2 and tyrosinase |TYR) with a high degree of selectivity for the compounds; two of the inhibitors were highly selective to the target protein (GZMB and TYR). These leads can be rapidly taken to animal models78,79 for efficacy and toxicity testing and may provide a rationale for pharmaceutical drug development. As a backup, 363 bioactive compounds were discovered in the study. These compounds expand the drug discovery pipeline for the treatment of Vitiligo. The results provide further support for the concept of accelerated drug discovery approaches starting from the GWAS data by using chemoinformatics approaches. While a cure for Vitiligo may be far away, it is possible to develop a rationale for repurposing drugs currently approved for other indications for the treatment of Vitiligo. While previously the pharmaceutical industry was reluctant to take this approach, it is becoming increasingly attractive. Such repurposing is applicable not only for already approved drugs, but also for failed drugs.67 The great advantage of such an approach is that the toxicity, side effects and bioavailability information is already available from the FDA database. The genes associated with Vitiligo were found to be associated with numerous approved drugs and nutraceuticals. These drugs are linked to treatment of cancer, inflammation, autoimmune diseases, immunosuppression, infections, arthritis, transplant rejection and skin disorders including psoriasis. Thus, these drugs can readily be tested in mouse models and in the clinic either as a single agent or in combination with other treatments such as phototherapy or Excimer laser. At the very least, these drugs could help fill the void that currently exists for the treatment of Vitiligo.
The Genome Wide Association datasets provided an attractive starting point for gene target discovery for the skin disease Vitiligo. Use of chemoinformatics and chemogenomics approaches led to the identification of druggable targets, bioactive compounds, FDA approved drugs and nutraceuticals associated with the Vitiligo genome. In view of the serious shortage of drugs for Vitiligo, which affects over 1% of the global population, the drugs identified in this study can be easily repurposed for the treatment of Vitiligo.
This work was supported in part by the Genomics of Cancer Fund, Florida Atlantic University Foundation. I thank the CanSar gene annotation tool for valuable datasets. I also thank Jeanine Narayanan for editorial assistance.
The author declares no conflict of interest.

©2015 Narayanan. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.