MOJ
eISSN: 2373-4442


Research Article Volume 4 Issue 3
1Molecular and Cytogenetics Department, Apollo Hospitals, Jubilee Hills, Hyderabad, Telangana, India
2Department of Clinical Laboratory Sciences, College of Applied Medical Sciences, King Saud University, Riyadh-, Saudi Arabi
Correspondence: Kavitha Matam, Molecular Biology and Cytogenetics Department, Apollo Health City Building, Apollo Hospitals, Jubilee Hills, Hyderabad, Telangana, India
Received: January 01, 1971 | Published: November 25, 2016
Citation: Matam K, Goud I, Vempati R, Khan IA (2016) Screening of Glioma Patients for 1p/19q Region with Fluorescent Probes. MOJ Immunol 4(3): 00132. DOI: 10.15406/moji.2016.04.00132
Oligodendroglial tumors represent approximately 4-7% of all gliomas. The discovery of 1p and 19q chromosomal arms deletion in glial tumour influence the diagnosis and more accurate prediction of chemotherapy response. As a result, an attempt has been made to detect deletion using fluorescent insitu hybridization (FISH) and to determine its prognostic value in a cohort of glial tumour patients from Hyderabad population. The FISH analysis was carried out on 90 FFPE tissue sections by using Vysis LSI 1p36/LSI 1q25 and LSI 19p13/LSI 19q13 dual colored FISH probe sets. Signals were scored from 150-250 non-overlapping, intact nuclei. The analysis for 1p and 19q deletions was observed in (21/35) 60% of oligodendroglyomas which included (8/21) 38.1% of grade II and (13/21) 61.9% of grade III. Isolated 19q deletion was seen in (1/21) 4.7% & lone 1p loss was not observed in oligodendroglyomas. In mixed oligoastrocytomas combined 1p/19q loss was observed in (7/16) 43.75% cases, including one grade II and 6 grade III tumors and 1/16 (6.25%) showed isolated 1p loss & 19q deletion. This disorder was not observed in astrocytomas. The oligodendroglial phenotype was found to be significantly associated with a loss of 1p (p< 0.05), a loss of 19q (p<0.05) and a combined loss of 1p and 19q (p< 0.05). Frontal location of a tumor occurred to be a statistically significant factor unfavorable for prognosis, p<0.05. The result of our study concludes 1p/19q deletions have prognostic significance and determines deletion 1p/19q by FISH into diagnostic and treatment algorithm in gliomas.
Keywords: gliomas, 1p/1q, fluorescent insitu hybridization, oligodendroglial tumors, fluorescent probes, ependymomas, fish
Gliomas are common in adult brain tumors and specific forms of gliomas such as ependymomas, astrocytomas (grade I-IV), oligodendroglyomas (grade II-III) and mixed oligoastrocytomas (grade II-III) have been varied in different grades as per world health organization criteria.1 Environmental risk factor for glioma is exposure to moderate to high doses of ionizing radiation and etiology of glioma genetic component contains an elevated glioma risk among individuals with a family history of glioma. The advanced understanding of aetiology of glioma has potential to facilitate treatment development, and thereby to improve the outcome of the disease.2 The pathogenesis glioma remains largely unknown.3 The diagnosed patient with glioblastoma has the median survival time for 14 months after the improved expansion of novel drugs and therapeutics such as chemotherapy, radiotherapy and tumorectomy.4 Gliomas are common in age of onset, gender in male, white race and non-Hispanic ethnicity.5 The identification of oligodendroglioma and astrocytoma in tissue sections can be performed with single-cell level consist of TP53, ATRX, IDH1R132H mutations and in situ hybridization for 1p/19q providing information on chromosome copy numbers.6 Reifenberger et al.7 has reported that allelic loss of chromosome 19q13 was detectable in 81% oligodendroglial tumors, ~ 75% also exhibited Loss of Heterogeneity at 1p36 locus. Oligodendroglial tumor patients has better prognosis than astrocytic neoplasms and patients with tumor contains 1p/19q co-deletion appear to be peculiarly sensitive to treatment.8 The deletions of 1p/19q chromosomes are associated with the genotype and phenotype of oligodendrogliomas.9 There are no specific studies have been carried out in our population and therefore we made an attempt to carry out the hospital based study in glioma patients in south Indian population for detecting the deletions in 1p/19r region of chromosomes by fluorescent insitu hybridization.
The initial step for this study was performed with the ethical approval from Apollo hospitals, Hyderabad, India. Written informed consent was approved from all the patients’ participated in this study and this is one of our inclusion criteria. The pathologist was identified the Glioma patients based on the WHO criteria. In this study, we have recruited the 90 tumor specimens, selected from department of pathology, Apollo hospitals, Hyderabad, India during the study period of January 2010-December 2013. We have collected 25 paraffin blocks oligodendrogliomas and 35 blocks for mixed oilgoastrocytic tumors. The hematoxylin and eosin slides were cross verified by senior pathologist based on WHO criteria. Fluorescence insitu hybridization (FISH) analysis was performed with dual color probes (LSI 1p36/LSI 1q25 and LSI 19p13/LSI 19q13) in paraffin embedded blocks prepared slides from the selected tumors. Dual probe hybridization was performed with the initial set of probe consist of spectrum orange color for locus 1p36 and spectrum green color for the control locus 1q25, whereas, the second probe will be appearing at spectrum orange at locus 19q13, and spectrum green for control locus. The whole FISH analysis was performed at the department of cytogenetics and molecular biology, Apollo hospitals as per the Li et al.10 studies. From each slide, 200 interphase non-overlapping nuclei signals were scored. Overall, 4 signals (2 red and 2 green) were detected in normal diploid nuclei and minimum of 50% and above nuclei had showed one signal has scored the deletion.
SPSS statistical software was used for the statistical analysis for Chi square test. A P-value of 0.05 was considered as statistical significance level.
One hundred and nineteen glial tumors were diagnosed in the Department of Pathology between January 2010 and December 2013.Of these, 58(48.7%) were Ass, 10(8.44%) are ependymomas. The pure OGs are 35(29.4%) and MOAs comprised 16(13.4%). All were tumor excision specimens. Fouty one patients were males (80.3%) and 10 (19.6%) patients were females. The patients’ characteristics are summarized in Table 1. Of the 35 cases of pure OG included in the study, 23 (65.7%) were grade II and 12 (34.2%) grade III. Of the 16 MOAs, 11 (68.75%) were grade II, 5 (31.25%) grade III; and amongst the 15 ASs (25.8%), 11 (73.3%) were grade II and 4 (26.6%) grade III. Fifty tumors occurred in adults (more than 18 years of age). There were 2 pediatric cases, 1 AS Grade II, 1 OG grade II (Table 1). A male preponderance was seen in all the histological types. Most of the tumors were frontal in location followed by temporal (Table 2). Rest involved other sites such as parietal lobe, diencephalon. Thirty-four tumors (66.6%) were centered in the frontal lobe, 12 (23.5%) in the temporal lobe, 3 (5.8%) in the parietal lobe, and 2 (3.9%) in diencephalon; no tumors were in the occipital lobe. Tumors in the frontal lobe showed 1p/19q loss in 76.4% of cases, whereas 83.3% of tumors in the temporal lobe had maintenance of 1p/19q. All three tumors in the Parietal and 2 in diencephalon showed preservation of 1p/19q. There was a significant association between frontal lobe location and loss of 1p/19q and temporal lobe and maintenance of 1p/19q (P< 0.05) (Table 2).
|
Diagnosis |
N (%) |
Age (Years) |
Gender (M:F) |
Tumor location (n) |
|
AS II |
14 (15.5%) |
10-49 |
10:4 |
F=11, T=2, P=1, D=0 |
|
AS III |
6 (6.7%) |
32-50 |
6:0 |
F=5, T=1, P=0, D=0 |
|
OG II |
33 (36.7%) |
25-62 |
21:12 |
F=24, T=5, P=3, D=1 |
|
OG III |
13 (14.4%) |
19-74 |
8:5 |
F=11, T=2, P=0, D=0 |
|
MOA II |
17 (18.9%) |
29-51 |
13:4 |
F=13, T=3, P=1, D=0 |
|
MOA III |
07 (7.8%) |
34-58 |
6:1 |
F=5, T=1, P=1, D=0 |
Table 1 Diagnosed tumors as per the grades
*M=male, F=Female, F= Frontal, T=Temporal, P=Parietal, D=Diencephalon
|
Tumor region |
Loss of 1p/19q (n%) |
OGs & MOA with 1p/19q maintenance |
p value |
|
Frontal |
26 |
8 |
<0.05* |
|
Temporal |
2 |
10 |
<0.05* |
|
Parietal |
0 |
3 |
- |
|
Diencephalon |
0 |
3 |
- |
Table 2 Location of Tumor with 1p/19q status
Fluorescence in situ hybridization assay
Majority (60%) of OGs harbored combined 1p/19q loss (21/35) (Figure 1). These included 38% of grade II (8/21) and 61.9% of grade III OGs (13/21). Isolated 19q deletion was seen in 4.76% of OGs (1/21), which on histomorphology belonged to WHO grade II (Figure 2). Lone 1p loss was not found in any of the OGs. Amongst the MOAs, combined 1p/19q loss was observed in 43.75% cases (7/16), including one grade II and 6 grade III tumors. Approximately 6.25% showed isolated 1p loss (1/16). A similar percentage showed isolated 19q loss (Figure 2). Both these were grade II tumors. None of the grade III MOAs harbored isolated chromosomal abnormalities. In contrast to OGs and MOAs, none of the ASs showed loss of 1p and/or 19q which included 2 pediatric cases (Figure 1). None of the five control cases showed either combined or isolated 1p/19q loss (Table 3). On statistical analysis, the oligodendroglial phenotype was found to be significantly associated with a loss of 1p (p< 0.05), a loss of 19q (p< 0.05) and a combined loss of 1p and 19q (p< 0.05).
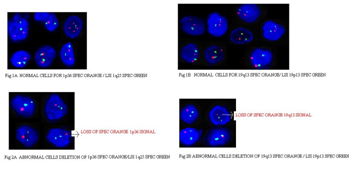
Figure legends
Figure 1A Normal cells for 1p36 spec orange/LSI 1q25 spec green.
Figure 1B Normal cells for 19q13 spec orange/LSI 19p13 spec green.
Figure 2A Abnormal cells Deletion of 1p36 spec orange/LSI 1q25 spec green.
Figure 2B Abnormal cells deletion of 19q13 spec orange/LSI 19p13 spec green.
|
|
Combined1p/19q loss (%) |
Isolated 1p loss (%) |
Isolated 19q loss (%) |
|
Oligo dendroglyoma |
21(60%)11 |
0 |
1(4.76%) |
|
Mixed Oligoastrocytoma |
7(43.75%)6 |
1(6.25%) 1 |
1(6.25%) |
|
Astrocytoma |
0 |
0 |
0 |
|
Adult n= 13 |
0 |
0 |
0 |
|
Pediatric n = 2 |
0 |
0 |
0 |
|
Normal Brain Parenchyma |
0 |
0 |
0 |
Table 3 1p/19q status in different Oligodendrogliomas
The molecular-cytogenetic study on chromosome 1p and 19q status has become an essential step in the treatment of oligodendroglial tumors. Codeletion of 1p and 19q whole arms is strongly correlated with a better response to standard treatment with radiotherapy and chemotherapy as well as a better overall survival. Earlier studies on molecular techniques are described in the literature to study the chromosomal status of tumor cells, including FISH, polymerase chain reaction, quantitative microsatellite analysis, loss of heterozygosity by microsatellite analysis, comparative genomic hybridization array and multiplex ligation - dependent probe. All these techniques have their advantages and disadvantages but the most widely used among them is FISH because it can be performed by fluorescent microscopy on paraffin embedded tumor tissue sections and is thus easily accessible to most pathology laboratories.11
The co-deletions in chromosome 1p and 19q regions has been identified in 50-90% of oligodendrogliomas.1 and in our study we have identified 20% co-deletions. Earlier studies have performed the whole genome sequence studies and identified the mutations on chromosome 19p13.2 and 1p31.1 in oligodendrogliomas.12 Based on these data, not surprisingly, many medical centers request molecular testing for chromosome 1p and 19q status by FISH on gliomas since their pathological evaluation & subtyping relies mainly on the light microscopic appearance, Although the histomorphologic criteria for their diagnoses are well established, a significant proportion of cases pose the diagnostic challenge. In this regard, the current study was performed to analyse the frequency of 1p/19q deletion, in various subtypes and grades of glial tumors cohorts from Telangana region. FISH has been shown a reliable method for the analysis of the 1p 19q status in oligodendroglial tumors.13 For the 51 subjects with Oligodendrogliomas & MOAs this study, the median age at diagnosis was 44 years (range, 19-74 years), and there was no difference in age between patients with maintenance or loss of 1p/19q. 80.3% patients were males and 19.6% patients were females. In the subgroup of patients with concurrent 1p/19q loss, there was a male predominance (78% versus 22%), whereas the subgroup of patients with 1p/19q maintenance showed a slight female predominance (56% versus 44%); however, differences were not significant. Our study indicated a significant association between frontal lobe location and loss of 1p/19q and temporal lobe and maintenance of 1p/19q (P < 0.05*) (Table 2), similar to the other studies.14 We have found that tumors developing in the frontal lobe were of a much higher incidence of 1p/19q loss. Loss of 1p/19q is considered a strong prognostic factor reported to occur in 50 – 90% of oligodendroglial tumors and around 27-50% of oligoastrocytomas and is comparatively lower (8 to 25%) in Ass.15-19 Our results in OG tumors showed combined 1p/19q loss in 60% which included 38% of grade II and 61.9% of grade III OGs. In MOAs, combined 1p/19q loss was observed in 43.75% cases, including 14.28% of grade II and 85.7% of grade III tumors (Table 3) which is in concordance with the above studies. None of the 15 cases of ASs showed deletion of either 1p or 19q. The oligodendroglial phenotype was highly associated with isolated or combined 1p/19q loss (P <0.05*) (Table 3). To the best of our knowledge, this is the first report on glial tumor patient’s prevalence rates by FISH from Telangana population. Reddy et al.18 studied 77 paraffin-embedded tissues using FISH for detecting 1p/19q loss. Combined 1p/19q deletion was found in 65% OGs, 60% MOAs and only 25% of ASs in his study.
Loss of 1p and 19q chromosomal arms is one of the most important molecular signatures in the diagnosis of central nervous system tumors. It is quite specific to oligodendroglial tumors and due to lack of any other phenotype and genetic marker typical of these neoplasms, it becomes an interest for pathologists. Cairncross et al.20 examined 39 anaplastic oligodendroglioma patients, 37 of whom had received PCV chemotherapy. Allelic loss of 1p was a statistically significant predictor of chemosensitivity, and combined loss of 1p and 19q was significantly associated with both chemosensitivity and longer recurrence-free survival following chemotherapy. Moreover, Smith et al.21 have demonstrated that the association of 1p and 19q loss with prolonged survival is also evident in low-grade oligodendroglioma patients and that this association may be independent of PCV chemotherapy. Ręcławowicz1 et al.22 in his study proved that combined loss involving chromosomes 1p and 19q is a strong factor for better prognosis.
As we showed and this observation supports earlier publications, deletions of chromosomal arms 1p and 19q are mutually strongly correlated. Exclusive 19q deletion was found in (1/21) 4.76% of OGs grade II. Similarly, isolated deletion of 1p, 19q is seen in 6.25% (1/16) of MOAs. Lone 1p loss was not found in any of the OGs (Table 2). We have not detected isolated deletions of either of these loci or the combined deletions in astrocytomas which are consistent with the results of previous surveys.
Since not all oligodendrogliomas demonstrate 1p/19q deletion, morphological features that could help differentiate these two groups were deeply sought. Therefore, we may conclude that when the tumour presents unequivocal morphological features of oligodendroglioma, 1p/19q deletion may be suspected. Pathology, radiology and cytogenetic studies play a complementary role in diagnosis, decision making and treatment.
Among presumed predictive factors in our study, three turned out to be statistically significant: histological type, 1p/19q deletion and localization (frontal vs. other). These results are in concordance with literature data, which confirm strong influence of the first two factors on prognosis.
Together, these data demonstrate the utility of 1p/19q status as a diagnostic marker in selected cases. Additionally, it can be considered as a point of reference with which to measure diagnostic criteria. Since a major goal of classification of grade 2 to 3 gliomas is to stratify patients into clinically distinct groups, it would be prudent to ensure that the majority of cases diagnosed as oligodendroglioma in daily practice exhibit combined 1p/19q loss because this signature is associated with both classic oligodendroglial features and improved outcome.
Detection of 1p/19q deletion in OGs & MOAs has potential benefits. Firstly, due to its high incidence in oligodendrogliomas, it has a huge value as an additional tool in their correct and objective diagnosis. In that regard, it can serve as an adjunct to histology and may serve as a point of reference for glioma diagnosis. Secondly, the inclusion of a patient in a specific prognostic group may influence the intensity of conducted treatment. The above features constituted the basis to include FISH analysis to detect these genetic alterations in patients with gliomas because of its higher resolution & higher sensitivity. Results of the presented paper support application of 1p/19 detection in common neurosurgical and pathological practice.
None.
None.

©2016 Matam, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.