MOJ
eISSN: 2373-4442


Mini Review Volume 5 Issue 5
1Department of Biology, University of Mustapha Stambouli, Mascara, Algeria
2Department of Biology, University of Oran (Es-senia), Oran, Algeria
Correspondence: Amina Meliani, Department of biology, University of Mustapha Stambouli, Mascara, Avenue Cheikh El Khaldi, Mascara 29000, Algeria, Tel 002-135-518-058-73
Received: January 01, 1971 | Published: May 18, 2017
Citation: Meliani A, Bensoltane A (2017) Pseudomonas Chemotaxis, Motility and Host-Pathogen Interactions. MOJ Immunol 5(5): 00167. DOI: 10.15406/moji.2017.05.00167
Infection with the fluorescent pathogen Pseudomonas aeruginosa leads to gastrointestinal infections, dermatitis, bacteremia and a variety of systemic infections. Thus, within a very complex chemosensory system this bacterium has requires an adaptive strategy to escape to the immune system. It chemosensory system has attracted a significant interest because of the very complex molecular diversity of this one (> 20 chemotaxis (che) genes). With this diversified chemotaxis system, this bacteria moves from cell to cell by a twitching motility and respond in a behavioral manner. For this, it can be viewed as an important prelude to infections and serious clinical challenge.
Keywords: Pseudomonas aeruginosa, infections, chemotaxis, twitching
Bacterial chemotaxis is a biased movement towards higher concentrations of life-sustaining nutrients and lower concentrations of toxins. It involves sensing a gradient of chemicals as small as a few molecules.1 Furthermore, this movement and under the influence of a chemical gradient, either toward (positive chemotaxis) or away (negative chemotaxis) from the gradient helps bacteria to find optimum conditions for their growth and survival.2 Motile bacteria have the ability to sense changes in the concentration of chemicals in environments and respond to them by altering their pattern of motility. This behavioral response is called chemotaxis. Chemotaxis signaling pathways control flagellar motility by regulating the frequency at which the flagellar motor changes its direction of rotation or the speed at which the flagellar motor rotates. This mode of control is conserved across flagellated bacteria, regardless of flagellar arrangement or number.1 Thus, chemotaxis signaling also controls twitching, the movement of cells on moist surfaces mediated by type IV pili (TFP), but the mechanisms involved are distinct from those controlling flagellum-dependent chemotaxis.3
The Pseudomonads also show chemotactic responses to various chemical compounds, including amino acids, organic acids, sugars, aromatic compounds and inorganic ions.4 Thus, many aspects of chemotaxis are now understood, at least superficially, but many questions remain especially in the case of Pseudomonas aeruginosa. This bacterium can modulate the immune response, reminiscent of helminth parasites, and antibiotic resistance due to the production of extracellular enzymes (e.g. β-lactamase).5 In humans, Pseudomonas aeruginosa infections tend to occur in association with epithelial cell damage to the skin or eye or medical devices such as catheters or ventilators or in immune-compromised individuals. In addition to these illnesses, P. aeruginosa lung infections are common in individuals with chronic obstructive pulmonary disease (COPD), ventilator-associated pneumonia (VAP), and cystic fibrosis (CF).6
Furthermore, the outcome of infections and establishment of disease depends on both host defense and bacterial capacities. The latter include its autonomic efficiency to grow, divide, and adapt to the environment, and the ability to sense, and communicate with their neighbors in the population to accomplish cooperative activities, e.g. biofilm formation and production of virulence factors.7 Consequently, elucidating the motility and chemotactic mechanisms for Pseudomonas spp. can be beneficial in many studies extending to bioremediation and host-pathogen interactions.8 Low permeability of its outer membrane by a complex set of efflux pump systems and secretion of alginate during biofilm formation are major factors that allow the pathogen to become highly virulent and resistant to multiple antibiotic agents. Adding to these factors, other bacterial exoproducts such as lipopolysaccharides and elastase induce harmful pathogenesis resulting in tissue destruction.9
In recent years, chemotactic responses studies between bacteria and self have contributed to a more informed view of the adaptative mechanisms used by P.aeruginosa. In this bacterium, contact is mediated by several adhesins, particularly type IV pili (TFP), long motorized fimbriae that also provide cells with surface-specific twitching motility and are essential to virulence and biofilm formation.11 Successive TFP extension, attachment, and retraction promote intimate association with surfaces and motility along them. Because TFP dynamically interact with the substrate, they mechanically couple cells with surfaces. Consequently, although TFP have been viewed as adhesion and motility structures, TFP could also potentially function as mechanical sensors to rapidly signal surface contact.12 This mini review provides some insight on the P.aeruginosa chemotaxis and twitching motility.
Pseudomonas aeruginosa Virulence and Cyclic AMP
Many virulence factors associated with P.aeruginosa infection (Figure 1) are regulated by the small molecule second messenger adenosine 3’, 5’-cyclic monophosphate (cAMP or cyclic AMP).13 In the case of P.aeruginosa, this messenger is believed to control gene expression through allosteric regulation of the transcription factor Vfr (Virulence factor regulator), which is a member of the cAMP receptor protein (CRP) family.14 Thus, cyclic AMP and Vfr appear to be the central components controlling a global virulence gene response in P.aeruginosathrough regulation of multiple virulence systems including type IV pili (TFP),13,15 the type II secretion (T2S) system and secreted toxins,14,15 type III secretion (T3S),13 quorum sensing (QS)4 and flagellar biogenesis.16
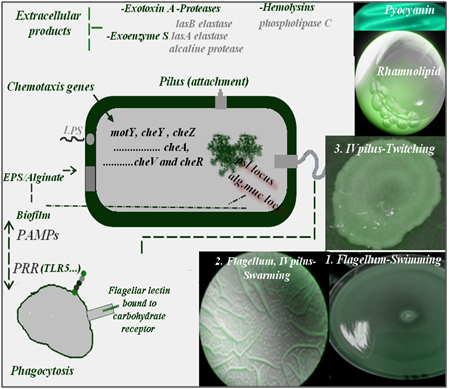
Figure 1 Biofilm formation and virulence are coupled in P.aeruginosa, since a variety of components such as flagellum and type IV pili play a fundamental role in P.aeruginosa biofilm formation and virulence. These thread like proteinaceous organelles display several modes of motility such as swimming, swarming and twitching motility and may play important roles in host–pathogen interactions. Note that TLR5 is a sensor for monomeric flagellin, known to be a virulence factor. Flagellin released from P.aeruginosa triggers airway epithelial TLR5 signaling NF-kB, and causing production and release of proinflammatory cytokines that recruit neutrophils to the infected region.
Beatson et al.15 reported thatthese factors are directly or indirectly controlled by the transcriptional regulator protein Vfr (virulence factor regulator). Vfr positively regulates production of exotoxin A (ETA or ToxA), type IV pili (Tfp), a type III secretion system (T3SS), and the las quorum-sensing system which, in turn, controls the expression of hundreds of additional genes, including multiple virulence factors.17 Interestingly,P.aeruginosa encodes two intracellular adenylate cyclases (CyaA and CyaB) responsible for cAMP synthesis.13 Nevertheless, mutants lacking both cyaA and cyaB exhibit reduced virulence factor expression and is severely attenuated in an adult mouse model of acute pneumonia.18 In addition, whole-genome expression profiling revealed that the transcriptomes of P.aeruginosa mutants defective in cAMP synthesis or lacking vfr are nearly identical, suggesting that Vfr activity is dependent on cAMP availability.19 It is also noteworthy, that Vfr is known to control twitching motility in P.aeruginosa. Another regulator FimL has been identified that affects twitching motility at least in part through modulation of Vfr production. FimL affects the regulation of type IVpilus assembly and function rather than production. While both fimL and vfr mutants show reduced levels of surface-assembled pili compared with wild-type, the defect is more severe in fimL mutants-an observation which supports the notion that FimL might also be controlling additional gene products necessary for functional type IV pili.4
Pseudomonas aeruginosa chemosensory system
It is obvious that genetic organization and physiological observations suggest that α proteobacteria and P. aeruginosa have different chemotaxis systems.20 P.aeruginosa has a very complex chemosensory system with more than 20 chemotaxis (che) genes in five distinct clusters and 26 mcp-like genes.21 The Chp system was previously implicated in the production and function of type IV pili (TFP).22 The Che and the Che2 systems, both homologous to the E. coli Che chemotaxis system, have been implicated in flagella-mediated chemotaxis,20 while genes in Pil-Chp cluster and Wsp cluster are involved in type IV pilus synthesis, twitching motility and biofilm formation, respectively.23 This bacterium also hasmultiple copies of E. coli-like chemotaxis genes arranged infive clusters.21 Two che clusters, cluster I and cluster V, which encode homologues of the six chegenes found in E.coli, have previously been shown to be essential for chemotaxis by P.aeruginosa.24 Cluster IV has been shown to be involved in twitching motility (Figure 2).25 Furthermore, nine P.aeruginosa MCPs have been identified as for amino acids, inorganic phosphate, oxygen, ethylene and volatile chlorinated aliphatic hydrocarbons, whereas three MCPs were demonstrated to be involved in biofilm formation and biosynthesis of type IV pilus.26 The Chp system was previously shown to control TFP production and twitching motility, Fulcher et al.21 analysis of TFP function also revealed that although twitching motility is ultimately dependent on TFP biogenesis, the Chp system exerts cAMP-independent regulatory control over TFP function. Currently the mechanism by which the Chp system regulates twitching motility is not known but one possibility is via regulation of TFP extension and retraction.
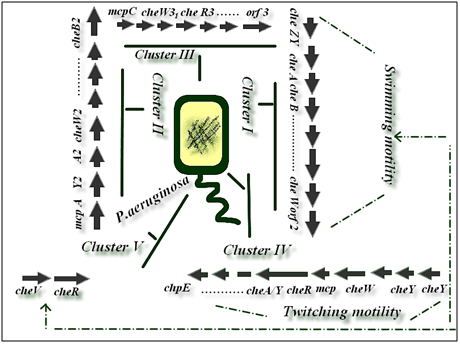
Figure 2 Bacterial species belonging to the genera Pseudomonas have been shown to possess multiple gene clusters involved in chemotaxis-like signaling pathways and other cellular functions. The biogenesis and function of type IV pili in P.aeruginos are controlled by more than 40 genes, including proteins involved in the structure, regulation of pilus assembly and twitching motility.4 Clusters I and V are involved in swimming motility chemotaxis but cluster IV is involved in twitching motility.20
Twitching motility and pathogenesis
Previous studies have shown that little is known of the mechanisms by which P.aeruginosa moves from cell to cell or by which it translocates (crosses) a multilayer epithelial barrier.27 Pseudomonas aeruginosa exhibits three types of motility flagellum-mediated swimming, flagellum and type IV pilus-mediated swarming, and type IV pilus-mediated twitching (Figure 2).4 The biogenesis and function of type IV pili in P.aeruginosa are controlled by more than 40 genes, including proteins involved in the structure, regulation of pilus assembly and twitching motility.4. As described above, in P.aeruginosa, twitching is controlled by a number of regulatory systems that sense external signals most of which are unknown and transduce them to modulate pilus extension and retraction. Both physical (e.g., viscosity) and chemical (e.g., phospholipids, iron) signals that influence twitching.12 P.aeruginosa and some other bacterial pathogens use twitching as a form of surface-associated motility that involves the extension, tethering, and retraction of polar type IV pili (T4P).28,29 Twitching is a mechanism for bacterial motility along a surface driven by type IV pili,29,30 micron-sized polymeric cell appendages that play a role not only in motility but also in cell-cell adhesion, cell-surface adhesion and horizontal gene transfer.31 Twitching motility is a mode of solid surface translocation that occurs under humid conditions on semisolid or solid surfaces, is dependent on the presence of retractile type IV pili, micron-sized polymeric cell appendages that play a role not only in motility but also in cell-cell adhesion, cell-surface adhesion and horizontal gene transfer.31 Furthermore, twitching motility is primarily a social form of movement. Twitching involves cell-cell interactions and movement along the long axis of the cells, with little to no movement being observed in isolated cells. Second, twitching results from a sequence of extension, tethering, and retraction of TFP.32
Twitching bacteria can also secrete slimes that are left as tracks on the surface traveled by a moving cell (Figure 3). The slimes comprise EPS and were shown to affect the activity of TFP in some bacteria.33 In addition, twitching bacteria often secrete slimes that can modify the surfaces on which cells move and thus further alter motility. Slime secretion might further alter the patterns and frequency of TFP binding-retraction and cause a progressive loss of motility. Furthermore, twitching motility plays a major role in both pathogenesis and biofilm formation. P.aeruginosa cells that bind to mucosal epithelial cells induce a variety of host cell signaling events and physiological responses.34 Type IV pili (T4P) are deployed in the early or acute phase of infection but are frequently lost owing to down regulation or mutation in chronic infections such as cystic fibrosis.35 It is interesting to speculate that type IV pilus-dependent surface motility directed by lipid effectors may be a critical event in pathogenesis in these and other organisms.25 Zolfaghar et al.36 data indicate that while twitching motility is not necessary for the induction of cytotoxicity or disease, it may play a role in advancing these processes by allowing the bacteria to spread through the tissue. Based on the present findings IV pili as flexible surface filaments are essential for the attachment of the pathogen to host epithelial tissues via also a twitching motility. According to O’Toole and Kolter37 twitching motility has been shown to be required for the initial attachment and development of a biofilm by P.aeruginosa. Thus, Rehm Bernd4 reported that the role of type IV pili in the pathogenesis of infection is still not settled. Many observations point towards a role of pilus in disease, for example Pilin deficient (pilA) mutants have decreased ability to damage epithelial cells, and have reduced cytotoxicity toward A549 and HeLa cells, and vaccination with purified pili can protect against infection with serologically related strains. Intratracheally but not subcutaneously, pili protein-immunized mice showed significant improvement of survival after intratracheal challenge with the P.aeruginosa PAO1 strain. P.aeruginosa pilT mutants are not infective in corneal tissue and exhibit reduced cytotoxicity to epithelial cells in culture.4
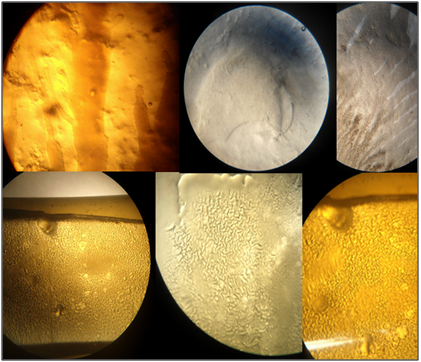
Figure 3 Variable twitching phenotypes displayed by different Pseudomonas aeruginosa. Swarming motility on 1.5 % agar Luria broth (LB) for 72h at 37±2°C. Note the secretion of slimes that are left as tracks on the surface traveled by a moving cell (discontinued circles).
According to pili drive motility through cycles of polymerization, adhesion and retraction: When attached to a surface, the retraction of a pilus into the cell body via its depolymerization pulls the bacterium forward along that surface.30 Twitching motility is independent of the presence of a flagellum. Surface translocation via twitching motility is powered by the extension and retraction of type IV pili and can manifest as a complex multicellular collective behavior that mediates the active expansion of colonies cultured on the surface of solidified nutrient media, and of interstitial colonies that are cultured at the interface between solidified nutrient media and an abiotic material.38
P.Aeruginosa via a vis innate immune response
To our knowledge, pili, flagella, exoenzyme S, and mucoid exopolysaccharide are recognized as major adhesins in P.aeruginosa. Invading pathogens are recognized by Toll-like receptors (TLRs) on epithelial cells and innate immunocytes, both of which are then activated to express inflammatory mediators. Thereafter, defense systems such as mucociliary clearance, phagocytosis and humoral immunity are promoted to neutralize the danger.10. Furtheremore, P.aeruginosa biofilms have various means to counterattack the immune defense in this review we highlight some aspect. Thus, chronic infections develop because the innate immune response is ineffective at clearing biofilm infections, irrespective of the location of the biofilm in the host.39 Within the innate immune response, phagocytic cells such as macrophages and polymorphonuclear leukocytes (PMNs) act as the first line of host defense.40. When analysing the interaction of neutrophils with P.aeruginosa biofilms generated in vitro, it was observed that neutrophils settled on biofilms, and they, however, did not move around and exhibited little or no bactericidal activity.41 Accordin to Tvenstrup Jensen, et al.42 P.aeruginosa biofilms downmodulated leukocyte functions. This modulation is regulated by the synthesis of rhamnolipids. These amphiphilic molecules are potential to fend off the leukocyte attack. Thus, in the interplay between biofilms and PMNs, rhamnolipids are a particularly important virulence factor. Jensen et al.43 showed that rhamnolipids produced by P.aeruginosacause PMNs to undergo necrotic death. Moreover, Alhede et al.44 and van Gennip45 showed that P.aeruginosa responds to the presence of PMNs by upregulating the synthesis of rhamnolipids. Eventually, the cytotoxic potential of rhamnolipids was linked to the pathogenicity of P.aeruginosa biofilms: rhamnolipids could actively fend off the neutrophils, leading to persistence of bacteria; moreover, lysed neutrophils may release their content of proteolytic enzymes, which may cause tissue damage, and hence progression of the inflammatory response.46
In this context it is noteworthy that flagellar motility had it part of this response. With regard to the contribution of bacterial motility to the recognition and clearance of P.aeruginosa, Amiel, et al..47 identified that bacterial flagellar motility is a pattern-recognition signal for phagocytic engulfment by innate immune cells. These authors identified that flagellar motility in P.aeruginosa is a critical phagocytic activation pattern both in vitro and in vivo. Loss of flagellar motility, independent of the flagellum itself, provides the bacteria with a ~100-fold increase in resistance to phagocytic uptake by macrophages, neutrophils, and dendritic cells. Lovewell, et al..48 revealed that the increase in resistance is due to bacterial activation of the host cell PI3K/Akt signaling pathway specifically by swimming motility. As flagellar swimming motility results in cellular Akt activation, which in turn regulates phagocytic recognition in the host cell and subsequent phagocytosis of the bacteria. Regardless, it is clear that the downregulation of flagellar motility, which is observed in certain infection such as chronic lung infections, directly contributes to P.aeruginosa persistence by providing the pathogen a potent means of phagocytic evasion. In summary, the data presented by Alarcon et al.49 showed that invasive P.aeruginosa can traverse multilayers of epithelial cell without disrupting (Transepithelial resistance) TER. Given that, and considering that invasive P.aeruginosa strains have the capacity to enter and exit cells, an intracellular pathway may be involved. Twitching motility, previously determined to be an important virulence factor for P.aeruginosa in multiple in vivo models, was found to be required for bacterial traversal in vitro and for bacterial exit from invaded cells, which was in turn reduced by a caspase inhibitor.50 Apoptosis has previously been shown to require twitching. According to these authors, one possible explanation for the contribution of twitching to virulence in vivo is that after bacteria adhere/invade, twitching facilitates the traversal of epithelial cell layers by enabling bacteria to exit the cells they have invaded through a process involving apoptosis. Thus, twitching motility in P.aeruginosa virulence in vivo remains to be determined.51
Most studies on Pseudomonas aeruginosa pathogenesis are focused on cell-associated and extracellular factors however the anthropocentric view of Pseudomonas twitching has distorted the scientists understanding of this pathogenesis and has attracted increased scientific attention. This review highlights how the chemotaxis system in Pseudomonas aeruginosa functions and how this system drives this bacterium to interact with the immune system and induce chronic inflammation. This can be achieved by understanding better the role of the IV pili as an important regulatory and virulence factor.
The anonymous reviewers are sincerely thanked for their beneficial suggestions to improve the manuscript.
The authors declare no conflicts of interest.

©2017 Meliani, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.