MOJ
eISSN: 2373-4442


Research Article Volume 5 Issue 7
1Department of Human Genetics, University of Pittsburgh School of Medicine, USA
2Departments of Dermatology, University of Pittsburgh School of Medicine, USA
3Departments of Immunology, University of Pittsburgh School of Medicine, USA
4Departments of Pathology, University of Pittsburgh School of Medicine, USA
5Departments of Bioengineering, University of Pittsburgh School of Medicine, USA
6Departments of Cancer, University of Pittsburgh Cancer Institute, USA
Correspondence: Walter J Storkus, Departments of Immunology, University of Pittsburgh, USA, , Tel 001-412-648-9981, Fax 001-412-383-5857
Received: January 01, 1971 | Published: October 23, 2017
Citation: Storkus WJ, Raveendran S, Fecek RJ, Brown MC, Fabian KLP et al. (2017) HSP90 Inhibition Enhances the Anti-Tumor Efficacy of Combination Chemo Immunotherapy Targeting DNA Repair Proteins. MOJ Immunol 5(7): 00181. DOI: 10.15406/moji.2017.05.00181
Although melanoma is generally considered to be resistant to genotoxic chemotherapeutic agents such as temozolomide (TMZ), we report that treatment of human and murine melanoma cell lines with TMZ up regulates their expression of DNA repair proteins (DNA‒RP) known to be stabilized by the molecular chaperone HSP90. Such TMZ‒induced changes in DNA‒RP expression were observed in vitro, as well as in vivo, in the B16 melanoma model. Based on our previous studies focused on the tumor‒associated HSP90 client protein EphA2 (a cell surface receptor tyrosine kinase), we hypothesized that TMZ‒induced DNA‒RP might serve as substrates for the generation of MHC class I‒presented peptide epitopes upon administration of the HSP90 inhibitor STA9090 (ganetespib). Notably, STA9090 promoted the degradation of TMZ‒induced DNA‒RP in melanoma cell lines in vitro, and the pre‒conditioning of progressively growing B16 melanomas in C57BL/6 mice with systemic TMZ + STA9090 treatment was found to sensitize melanoma cells to the anti‒tumor action of adoptively‒transferred DNA‒RP‒specific CD8+ T cells. The anti‒tumor efficacy of chemo immunotherapy combining TMZ, STA9090 and antigen‒specific adoptive T cell transfer was found to be associated with superior levels of Type‒1 CD8+ tumor‒infiltrating lymphocytes (TIL) in treated mice and more expansive areas of tissue apoptosis within the tumor microenvironment (TME). These data suggest possible translation of such combination chemo immunotherapy strategies into the clinic for the treatment of patients with melanoma or other forms of cancer.
Keywords: Adoptive cell therapy, CD8+ T cells, Chemotherapy, DNA repair proteins, HSP90 inhibitor, Immunotherapy, MHC class I, Temozolomide, Tumor microenvironment
ACT, Adoptive Cellular Therapy; ATM, Ataxia Telangiectasia Mutated Protein; ATR, ATM and Rad3‒Related Protein; BRAF, B‒Raf proto‒oncogene; BRCA, Breast Cancer; CFDA‒SE, Carboxy Fluorescein Diacetate Succinimidyl Ester; CFSE, Carboxy Fluorescein Succinimidyl Ester; CHEK, Checkpoint kinase; CM, Culture Medium; CTLA‒4, Cytotoxic T‒ Lymphocyte Associated protein 4; DC, Dendritic Cell; DNA‒RP, DNA Repair Proteins; DMSO, Dimethyl Sulfoxide; DOX, Doxorubicin; ELISA, Enzyme‒Linked Immuno Sorbent Assay; FMOC, Fluorenyl Methoxy Carbonyl; HDAC, Histone Deacetylase; HIP, Hsp70 Interacting Protein; HOP, Hsp70‒Hsp90 Organizing Protein; HSP, Heat Shock Protein; HSP90i, HSP90 Inhibitor; DSB, Double‒Strand Breaks; IFN, Interferon; IL, Interleukin; MACS, Magnetic‒Activated Cell Sorting; MART1, Melanoma Antigen Recognized by T cells 1; MHC, Major Histo Compatibility; MRE 11, Meiotic Recombination Protein 11; MRN, MRE11–RAD50–NBS1, NBN, Nibrin; PD1, Programmed Cell Death Protein 1; PD‒L1, Programmed Death‒Ligand 1; PRKDC, Protein kinase DNA‒activated Catalytic polypeptide; QPCR, Quantitative Polymerase Chain Reaction; SDS‒PAGE, Sodium Dodecyl Sulphate‒Polyacrylamide Gel Electrophoresis; TIL, Tumor‒Infiltrating Lymphocytes; TME, Tumor Microenvironment; TMZ, Temozolomide; TRP, Tyrosinase Related Protein; TUNEL, Terminal Deoxynucleotidyl Transferased UTP Nick End Labeling
Melanoma is the 5th most common form of cancer in the United States and is one of three cancer types that continue to increase in incidence.1 Metastatic melanoma, the most dangerous form of skin cancer, responds poorly to conventional chemotherapy, with a median overall survival of less than one year.2 Although the results of several recent clinical trials support the potent anti‒melanoma activity of immune checkpoint inhibitors (i.e. anti‒CTLA4, anti‒PD1 or anti‒PD‒L1 antibodies) as standard of care, the frequency of patients receiving durable treatment benefit remains limited.3‒6 Hence, there continues to be a clear need for the development of novel, effective (second‒line) therapy options for such patients.
Chemotherapeutic agents, such as temozolomide (TMZ), cause “in correctable” DNA lesions in treated tumor cells, most commonly DNA double‒strand breaks.7‒9 Tumor cells that are only moderately impacted (and not killed) by TMZ may amplify gene products associated with DNA repair to maintain cellular viability, leading to a state of acquired chemotherapy resistance.10 In the melanoma setting, DNA‒RP (such as ATR, MRN complex proteins (MRE11, Rad50, NBN), Rad51, PRKDC) are transcriptionally over expressed in a disease stage‒associated manner in association with chemotherapy‒resistance and poor overall survival.11‒18 Notably, tumor cell (over) expression of DNA‒RP may also be post‒translationally stabilized via the action of heat shock protein‒90 HSP90;11,13‒17,19 a molecular chaperone that is highly‒abundant in the cancer proteome.20 HSP90 forms the core of a super‒chaperone machine consisting of HSP70, HSP40, HIP and HOP, which extends the molecular lifespan of a growing list of client proteins, including signaling protein kinases, transcription factors, DNA‒RP and other cytosolic or nuclear proteins in their functionally mature and active conformations.19
While most targeted therapy agents antagonize 1‒2 individual proteins or signaling pathways, which might facilitate the rapid selection of compensatory pro‒oncogenic pathways in adapting tumor cells, HSP90 inhibitors (HSP90i) would be expected to coordinately disrupt multiple DNA‒RP clients (and a range of pro‒tumor signaling pathways) that might otherwise support the survival/progression of heterogeneous subpopulations of cancer cells.20 HSP90i are known to promote the conditional degradation of client proteins via the proteasome pathway,11,19,21 a major conduit for the generation of immunogenic peptides presented by MHC class I molecules for recognition by CD8+ T effector cells.22 Hence, we reasoned that melanoma cells conditioned by genotoxic chemotherapy agents to up regulate levels of DNA‒RP protein expression, and then treated with HSP90i to foster conversion of DNA‒RP proteins into peptide epitopes, might enhance tumor cell recognition by (therapeutic) DNA‒RP‒specific CD8+ T cells in vitro and in vivo.
Our data developed in both murine and human melanoma models indeed suggests that tumor expression of DNA‒RP may be upregulated in the presence of TMZ, and that at least a subset of tumor over expressed DNA‒RP may then be induced to undergo degradation after treatment with the HSP90i STA9090, in association with enhanced recognition of treated tumor cells by CD8+ T cells primed against DNA‒RP‒derived peptide epitopes in vitro. Furthermore, TMZ + STA9090 pre‒conditioning of established B16 melanomas in vivo enhances the anti‒tumor efficacy of adoptive cell transfer (ACT) using (anti‒DNA‒RP) CD8+ T cells. The anti‒tumor benefit of this combination (TMZ + HSP90i + ACT) chemo immunotherapy was associated with enhanced Type‒1 T cell infiltration into tumor lesions, with higher levels of apoptosis observed in the TME, and with the upregulated expression of the inflammation‒responsive PD‒L1 immune checkpoint molecule by tumor/stromal cells post‒treatment. Overall, these studies serve as a foundation for the design and implementation of novel chemo‒immunotherapeutic strategies for translation into the clinic for the treatment of patients with advanced‒stage melanoma or alternate cancer types.
Mice
Six‒ to 10‒week‒old female C57BL/6 (H‒2b mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and maintained in a pathogen‒free animal facility at the University of Pittsburgh. All animal experiments were performed in accordance with a University of Pittsburgh Institutional Animal Care and Use Committee (IACUC)‒approved protocol.
Tumor cell lines
The B16 murine melanoma (H‒2b) cell line was purchased from the American Type Culture Collection (ATCC; Manassas, VA). The murine BRAFV600EPTEN‒/‒ melanoma cell line BP23 was kindly provided by Dr. Jennifer Wargo (M.D. Anderson Cancer Center, Houston, TX) and then selected for resistance to the BRAF inhibitor dabrafenib (20 mM) for 1 month in vitro, yielding the BPR cell line, which established and progressed more uniformly than the parental tumor cell line after s.c. implantation in C57BL/6 mice (Fecek et al., unpublished results). The human melanoma cell lines, MEL526 and MEL624 were the kind gifts of Dr. Steven Rosenberg (National Cancer Institute, USA) and have been described previously.24 The EL4 (H‒2b; ATCC) thymoma was used as a negative control target for CD8+ T cell recognition assays. All tumor cell lines were cultured in complete media (CM; RPMI 1640 media supplemented with 100 units/mL penicillin, 100 μg/mL streptomycin, 10 mmol/L L‒glutamine, and 10% heat‒inactivated fetal bovine serum; all purchased from Life Technologies, Grand Island, NY) in a humidified incubator at 37 °C and 5% CO2. All cell lines were negative for known mouse pathogens, including Mycoplasma.
Western blotting
Melanoma cell lines were grown to 80–90% confluence and then incubated with Temozolomide (50 mM, Selleck Chem, Houston, TX) for 12h, in the absence or presence of the HSP90i STA9090 (50 mM; Synta Pharmaceuticals, Lexington, MA) for an additional 12h. Alternatively, animals were euthanized, and progressively‒growing tumors from control or TMZ +/‒ HSP90i treated tumor‒bearing mice were isolated by surgical dissection. Harvested cells/tissues were then incubated with lysis buffer, and cell‒free lysates resolved by SDS‒PAGE prior to electro‒transfer onto polyvinylidene difluoride membranes as previously described.25 Blots were probed with specific anti‒DNA‒RP (ATR, PRKDC, MRE11, NBN, Rad50, Rad51) or b‒actin pAb antibodies (all from Abcam, Cambridge, UK). Blots were imaged for 5 minutes on Kodak X‒Omat Blue XB‒1 film (Eastman Kodak, Rochester, NY) after incubation with horseradish peroxidase (HRP)‒conjugated goat anti‒rabbit Ig or goat anti‒mouse Ig antibodies (Santa Cruz Biotechnology) and the Western Lighting chemiluminescence detection kit (Perkin‒Elmer, Waltham, MA).
Quantitative real time polymerase chain reaction (qPCR)
Total RNA was extracted from either treated melanoma cell lines or total resected tumors from treated mice using the RNeasy Plus Mini Kit (Qiagen) including the gDNA Eliminator column. The Nanodrop ND‒1000 (CellBio SpA, Milan, Italy) was used to check the quality and quantity of RNA. Total RNA (1ug) was reversed transcribed into cDNA using the High Capacity RNA to cDNA kit (Life Technologies) and the cDNA added to Fast SYBR® Green Master Mix (Life Technologies) and used for quantitative PCR. Reactions were performed on a Step One PlusTM Real‒Time PCR thermo cycler (Applied Biosystems) using the recommended cycling conditions. All mRNA expression levels were normalized to the expression of the cellular housekeeping gene product HPRT1. Primer sequences were selected using Primer‒BLAST Genbank and are listed in Table S1.
DNA‒RP |
Primer (forward 5’‒3’) |
Primer (Reverse) |
Amplicon Size (bp) |
ATR |
TTGGAAGGGCAGCAAAAGGA |
CTCCAGAGACGGATGCAGAC |
90 |
PRKDC |
GACAAGTGCAGAAATGGAAGCA |
CAGCCTGGCTTCAGAAGAGT |
88 |
MRE11 |
GCCCCACAGATCCACTTGAC |
TTCCTCTAACTGCATCTTTCTCCA |
96 |
NBN |
CGGCTCCAGGAGAACCATAC |
ATGCCACAGTTTTTCCTCCCA |
70 |
Rad50 |
GCGTGCGAAGTTTTGGGATA |
AATGATGGTCGTCTTCCCCG |
107 |
Rad51 |
GCTGATGAGTTTGGTGTCGC |
TTGGGATCTGCAGCGAACAT |
86 |
Table S1 Real‒time (qPCR) primers used to amplify DNA‒RP transcripts in this study
Selection of MHC class I‒presented DNA‒RP peptide epitopes
Specific DNA‒RP peptides used in this study are listed in Table S2. These species were selected based on ranked scores generated in a web‒based algorithm predictive of the ability of peptides to be produced from parental proteins by proteasomal processing combined with their ability to bind (and be presented by) either H‒2Db or H‒2Kb class I molecules. All peptides were synthesized using 9‒fluorenylmethoxycarbonyl (Fmoc) chemistry by the University of Pittsburgh Cancer Institute’s Peptide Synthesis Facility (a Shared Resource). Peptides were >96% pure based on high performance liquid chromatography profile and mass spectrometric analysis performed by the University of Pittsburgh Cancer Institute’s Protein Sequencing Facility (a Shared Resource).
DNA‒RP |
AA Position |
AA Sequence |
Predicted H2‒Kb or ‒Db Presentation |
ATR |
1390‒1397 |
LSFAYGLL |
Kb |
200‒208 |
MNVQNVEFI |
Db |
|
PRKDC |
1187‒1194 |
LFYKFVPL |
Kb |
620‒628 |
SALINLVEF |
Db |
|
MRE11 |
208‒216 |
NSWFNLFVI |
Db |
109‒116 |
VNYQDGNL |
Kb |
|
NBN |
355‒363 |
SAPVNMTTY |
Db |
273‒281 |
VGITNTQLI |
Kb |
|
Rad50 |
391‒399 |
RQIKNFHEL |
Db |
1180‒1187 |
RNYNYRVV |
Kb |
|
Rad51 |
58‒66 |
KELINIKGI |
Db |
212‒219 |
VESRYALL |
Kb |
Table S2 DNA‒RP‒derived peptides predicted to be processed by the proteasome and presented by H‒2b class I molecules
Source: Full‒length DNA‒RP sequences were queried on the web‒based Immune Epitope Database and Analysis Resource using its MHC‒I Processing Prediction algorithm. Peptides were then selected for synthesis and immunologic analyses based on predicted MHC I IC50 results.
Generation of DNA‒RP‒specific CD8+ T cells for adoptive transfer therapy
To generate Ag‒specific CD8+ T cells for adoptive cell transfer experiments, naïve C57BL/6 (tumor‒free) mice were vaccinated s.c. (right flank) on days ‒21, ‒14 and ‒7 with a vaccine consisting of syngenic bone marrow‒derived dendritic cells (DC) transduced with recombinant adenovirus encoding mIL‒12p70 (i.e. DC.IL12; known to promote T helper‒independent activation of Type‒1 CD8+ T (Tc1) cells as previously described;26 for 48h at 37 °C, that were pulsed for 4h at 37 °C with pools of peptides derived from DNA‒RP as indicated (with each individual peptide present at a concentration of 1 mM). One week after the 3rd vaccination (i.e. day 0), the animals were euthanized, spleens harvested and CD8+ splenic T cells (MACSTM‒selected; Miltenyi Biotec, Auburn, CA) obtained as previously described.26 T cell specificity was determined in IFN‒g ELISA against melanoma vs. control EL4 vs. DNA‒RP peptide‒pulsed EL4 target cells, as described above. For adoptive cell therapy experiments, CD8+ splenic T cells were fluorescently labeled using the Vybrant CFDA‒SE cell tracer kit (Invitrogen) for 15 mins at 37 °C, just prior to washing and i.v. injection (in PBS) into melanoma‒bearing mice.
Tumor establishment and therapy models
Tumors were established by injection of 1 x 105 B16 melanoma cells s.c. into the right flank of syngeneic C57BL/6 mice. For therapy models, tumors were allowed to establish and grow through day 6, at which time, animals were randomized into cohorts of 5 mice/group, with each cohort exhibiting a mean tumor size of approximately 40‒50 mm2. For standard chemotherapy experiments, mice were treated with i.p. injection of TMZ (50 mg/kg/day) in 100 ml DMSO (Sigma‒Aldrich, St. Louis, MO) or vehicle control (DMSO) on days 6 and 7. For combinational chemoimmunotherapy treatment, tumor‒bearing mice were treated with TMZ (i.p., 50 mg/kg/day in 100 ml DMSO) or vehicle control DMSO on days 6 and 7, STA9090 (i.p., 25 mg/kg/day in 100 ml DMSO) or vehicle control DMSO on days 8 and 15, and adoptive transfer of 1 x 106 anti‒DNA‒RP CD8+ T cells (i.v. in the tail vein, in PBS) or vehicle control PBS on days 9 and 16. Tumor size (in mm2) was determined as the product of orthogonal caliper measurements monitored every 3 to 4 days thereafter. Mice were euthanized when tumors became ulcerated or they reached a size of 400 mm2, in accordance with IACUC guidelines.
IFN‒g ELISA
For tumor recognition assays, splenic CD8+ T cells were co‒cultured with melanoma cells, negative control EL4 (H‒2b; thymoma) or EL4 cells pre‒pulsed (for 4h at 37 °C) with DNA‒RP‒derived peptides (1 mM each) for 48h, after which, cell‒free supernatants were harvested and assessed for mIFN‒g concentration using a specific ELISA (BD Biosciences) as previously described.26 Data are reported as mean ± SD of quadruplicate determinations.
Immuno fluorescence staining and imaging
Tumor tissue was processed and sectioned as previously reported,26 followed by immunofluorescence staining and microscopy. The following primary antibodies were used for staining sections:rat anti‒mouse CD3, rabbit anti‒mouse T‒bet (Santa Cruz Biotechnology, San Diego, CA). Secondary antibodies included:Cy3‒conjugated goat anti‒rat Ig and Alexa488‒conjugated goat anti‒rabbit Ig (both from Jackson Immuno Research, West Grove, PA). TUNEL staining for detection of apoptotic cells was performed using a cell death detection kit (Roche Diagnostics, Indianapolis, IN) per the manufacturer’s instructions. All tissue sections were then briefly incubated with the nuclear counter‒stain 4′, 6‒diamidino‒2‒phenylindole (DAPI; Sigma‒Aldrich). After washing, sections were then covered in Gelvatol (Monsanto, St. Louis, MO) and a cover slip applied. Slide images were acquired using an Olympus Provis microscope (Olympus America, Center Valley, PA). Isotype control and specific antibody images were taken using the same level of exposure on the channel settings. Metamorph (Molecular Devices, Sunnyvale, CA) software was used for labeled cell quantification.
Flow cytometry
Before the staining of cells, Fc receptors were blocked with an anti‒CD16/CD32 antibody (Becton Dickinson). Single‒cell suspensions were directly stained with APC‒conjugated anti‒CD8 (eBioscience) and Annexin V‒FITC from the apoptosis detection kit Staining kit (Abcam) per the manufacturer’s instructions. Fluorescently‒stained cells were assessed using an LSR II flow cytometer (Beckman Coulter), with data analyzed using FlowJo software (Tree Star, Inc.).
Statistical analyses
Comparisons between groups were performed using a two‒tailed Student’s t‒test or one‒way Analysis of Variance (ANOVA) with post‒hoc analysis, as indicated. All data were analyzed using Sigma Stat software (Systat Software, USA). Differences with a p‒value < 0.05 were considered as significant.
TMZ promotes upregulated expression of DNA‒RP in murine and human melanoma cell lines in vitro and in vivo
To initially determine whether the genotoxic chemotherapeutic agent TMZ could alter tumor cell expression of DNA‒RP, melanoma cell lines were incubated in vitro with TMZ for 48h. Using Western Blotting analyses, we observed that TMZ treatment resulted in the coordinate increased expression of known DNA‒RP (including ATR, PRKDC, MRE11, NBN, Rad50, Rad51) when compared with expression of the b‒actin control protein by both murine (B16, BPR) and human (MEL526, MEL624) melanoma cell lines (Figure 1A & Table S3). Although increased expression of multiple DNA‒RP was observed for each tumor cell line examined after treatment with TMZ, the profile of upregulated DNA‒RP was not identical across all cell lines (with only MRE11 uniformly upregulated in all 4 melanoma lines evaluated). To determine whether similar results could be obtained in vivo, C57BL/6 were implanted with B16 melanoma cells s.c. in the right flank and tumors allowed to establish and progress for 6 days. On days 6 and 7, tumor‒bearing mice were administered TMZ (50 mg/kg/day in DMSO) or vehicle control DMSO. As shown in (Figure 1B), TMZ‒based therapy slowed B16 melanoma growth for approximately 1 week, at which time, tumors developed drug‒resistance and re‒established control growth kinetics by day 19 of the experiment. Western Blotting analyses of day 19 harvested tumors revealed coordinate upregulated expression of multiple DNA‒RP (e.g. ATR, MRE11, NBN, RAD50, RAD51) in vivo in tumor‒bearing mice treated with TMZ versus the control cohort (Figure 1C). The profile of DNA‒RP upregulated in B16 melanoma cells by TMZ was identical in vitro and in vivo (Figure 1A&1C).
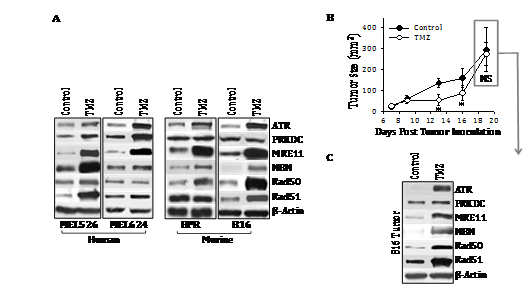
Figure 1 Treatment of melanoma cell lines with TMZ results in up regulated expression of a subset of DNA‒RP in vitro and in vivo. In A, human (MEL526, MEL624) and murine (BPR, B16) melanoma cell lines were cultured in the presence of TMZ (50 mM) for 3 hours at 37oC, at which time, tumor cells were lysed and SDS‒PAGE resolved proteins analyzed by Western Blotting using specific antibodies reactive against the indicated DNA‒RP or control b‒action (as described in Materials and Methods). In B, C57BL/6 mice bearing established s.c. B16 melanomas (day 7) were treated i.p. with TMZ (50 mg/kg in DMSO on days 7‒9) or with vehicle control. Tumor growth was monitored every 2‒4 days through day 19, with mean tumor size (+ SD) tumor size reported in mm2. In C, melanoma isolated from the tumor of median size in each cohort on day 19 was solubilized and subjected to Western Blotting analysis (Figure 1A). Specific DNA‒RP protein bands in blots were quantitated by densitometry scanning and normalized to control b‒action as described in Materials and Methods, with values reported in Table S3. Results are reflective of 3 independent experiments performed in each instance. *p < 0.05 vs. control; NS, not significant.
Melanoma Cell Line |
ATR |
PRKDC |
MRE11 |
NBN |
Rad50 |
Rad51 |
Figure 1A (In Vitro) |
||||||
MEL526 |
1.08 + 0.15 |
1.23 + 0.06* |
3.52 + 0.61* |
1.86 + 0.22* |
1.11 + 0.21 |
2.76 + 0.34* |
MEL624 |
1.66 + 0.25* |
1.38 + 0.15* |
2.45 + 0.31* |
1.04 + 0.10 |
0.99 + 0.03 |
1.01 + 0.18 |
BPR |
1.22 + 0.09* |
1.04 + 0.11 |
1.89 + 0.26* |
1.26 + 0.09* |
1.33 + 0.24 |
0.91 + 0.13 |
B16 |
1.62 + 0.18* |
0.95 + 0.13 |
4.05 + 0.37* |
1.51 + 0.16* |
2.41 + 0.28* |
1.39 + 0.21* |
Figure 1C (In Vivo) |
||||||
B16 |
4.45 + 1.21* |
1.04 + 0.06 |
3.12 + 0.63* |
2.44 + 0.29* |
1.84 + 0.30* |
1.51 + 0.31* |
Table S3 Densitometry analysis of Figure 1
Source:Western blots for (Figure 1A) and (Figure 1C) were analyzed by digital densitometry as outlined in Materials and Methods. Data were normalized to b‒Actin control protein expression in all cases. Reported values represent the ratio of normalized protein expression in TMZ‒treated tumor (cells)/untreated tumor (cells) for 3 independent experiments performed in each case. *p < 0.05 (gray highlights).
DNA‒RP expression in TMZ‒conditioned melanomas is subject to degradation in the presence of HSP90i
Since members of the DNA‒RP family of proteins have been previously suggested to represent HSP90 clients,13‒17 we next analyzed whether expression of DNA‒RP by TMZ‒conditioned B16 (murine) and MEL526 (human) cells was sensitive to treatment with the HSP90i STA9090 in vitro. As shown in (Figure 2&Table S4), we found that TMZ‒induced DNA‒RP expression was downregulated after co‒culture with STA9090 when compared with b‒actin protein controls. These data support the conclusion that at least a fraction of melanoma DNA‒RP undergo degradation when treated with HSP90i, theoretically providing a therapy‒associated source of MHC class I presented peptides allowing for enhanced CD8+ T cell recognition of treated melanoma cells.27
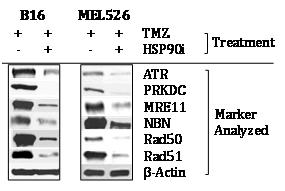
Figure 2 TMZ‒up regulated DNA‒RP in murine and human melanoma cell lines is degraded in the presence of HSP90i STA9090. Murine B16 and human MEL526 melanoma cells were pre‒treated with TMZ (50 mM) for 3h at 37oC, before subsequent culture in the absence or presence of HSP90i (50 mM) at 37oC for an additional 12h. Cells were then solubilized and DNA‒RP proteins analyzed by Western Blotting per (Figure 1A). Specific DNA‒RP protein bands in blots were quantitated by densitometry scanning and normalized to control b‒action as described in Materials and Methods, with values reported in (Table S4). Results are reflective of 3 independent experiments performed.
Melanoma Cell Line |
ATR |
PRKDC |
MRE11 |
NBN |
Rad50 |
Rad51 |
B16 |
0.33 + 0.12 |
0.14 + 0.11 |
0.19 + 0.07 |
0.64 + 0.17 |
0.20 + 0.13 |
0.25 + 0.18 |
MEL526 |
0.38 + 0.09 |
0.23 + 0.10 |
0.34 + 0.16 |
0.54 + 0.20 |
0.24 + 0.14 |
0.36 + 0.07 |
Table S4 Densitometry analysis of Figure 2
Source:Western blots for (Figure 2) were analyzed by digital densitometry as outlined in Materials and Methods. Data were normalized to b‒Actin control protein expression in all cases. Values represent the ratio of normalized protein expression in TMZ + HSP90i‒treated tumor (cells)/TMZ only treated tumor (cells) for 3 independent experiments performed in each case. All values with p < 0.05 vs. TMZ only control.
TMZ‒inducible DNA‒RP contain immunogenic peptides recognized by CD8+ T cells in wild‒type C57BL/6 mice
To determine the impact of TMZ and/or HSP90i on the generation of MHC I‒presented DNA‒RP‒derived peptide epitopes and consequent CD8+ T cell recognition, we first needed to identify immunogenic peptide species from DNA‒RP observed to be upregulated in melanoma after treatment with TMZ. To do so, we employed a web‒based algorithm that coordinately interrogates the likelihood that a peptide is processed by the proteasome, and that it can bind to either the H‒2Kb or H‒2Db class I molecules expressed in C57BL/6 mice. Based on these results, we synthesized 12 peptides (i.e. 2 top scoring peptides from each of the DNA‒RP ATR, PRKDC, MRE11, NBN, Rad50 and Rad51; (Table S2).
These peptides were then used to immunize naive C57BL/6 mice. Mice were vaccinated sub‒cutaneously a total of 3 times on a weekly basis with IL‒12 gene‒modified DC (which promote CD8+ T cell responses without the need for CD4+ T helper epitopes;26 pulsed with an equimolar pool of the synthetic DNA‒RP peptides (Figure 3A). Control mice were vaccinated with IL‒12 gene‒modified DC alone. One week after the final DC‒based vaccine, mice were euthanized and CD8+ T cells isolated from the spleen were cultured with the syngenic (H‒2b) antigen‒presenting cell line EL4, or with EL4 cells that had been pulsed in vitro with each of the individual DNA‒RP‒derived peptides used in the vaccine. As shown in (Figure 3B), mice vaccinated against DNA‒RP developed CD8+ T cells reactive against specific DNA‒RP peptides based on their IFN‒g secretion response quantitated by specific ELISA. Amongst the peptides (Table S2) analyzed, the Rad501180‒1187 peptide appeared most immunogenic in the pooled vaccine formulations, with the peptides ATR200‒208, PRKDC620‒628, MRE11109‒116, MRE11208‒216, NBN273‒281, Rad50391‒399, Rad5158‒66 and Rad51212‒219 also recognized to a lesser degree by vaccinated vs. control CD8+ T cells (Figure 3B). In contrast, the peptides ATR1390‒1397, PNA‒PKCs1187‒1194 and NBN355‒363 failed to elicit statistically‒significant, antigen‒specific CD8+ T cell responses in any vaccinated mice, and were considered non‒immunogenic in C57BL/6 mice.
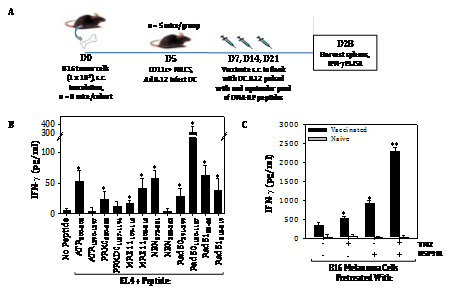
Figure 3 Vaccination against DNA‒RP‒derived peptides elicits specific CD8+ T cells in mice that preferentially recognize TMZ/HSP90i‒conditioned B16 melanoma cells in vitro. To generate antigen‒specific CD8+ T cells reactive against DNA‒RP, we first used a web‒based algorithm to select for those peptides most likely to be coordinately proteasome‒processed and MHC class I‒presented in the H‒2b C57BL/6 strain of mice (Table S2). Naïve mice were vaccinated s.c. a total of 3 times on a weekly regimen with syngenic DC.IL12 cells (i.e. DC engineered to produce mIL12p70 protein) that were loaded with an equimolar pool (1 mM for each peptide) of the chosen peptides (panel A schema, with additional detail in Materials and Methods). Splenic CD8+ T cells isolated from vaccinated or control mice were then co‒cultured at an effector‒to‒target cell ratio of 5:1 for 48h with EL4 (H‒2b, thymoma) cells, EL4 cells pulsed (for 4h at 37oC) with individual DNA‒RP peptides (1 mM, panel B), or against untreated B16 melanoma cells or B16 cells pre‒treated with TMZ +/‒ HSP90i (panel C). Cell‒free supernatants were then analyzed for mIFN‒g concentration using a cytokine‒specific ELISA. Results are reflective of 3 independent experiments performed in each instance. *p < 0.05 vs. EL4 or B16 control cells; **p < 0.05 versus all other cohorts.
Splenic T cells from mice vaccinated against pooled DNA‒RP‒derived peptides were also evaluated as effector cells against target cells including the negative control EL4 thymoma, untreated B16 melanoma cells, and B16 melanoma cells pre‒treated with TMZ (to upregulate DNA‒RP) +/‒ HSP90i (STAT9090, to conditionally promote MHC class I complex presentation of DNA‒RP‒degraded peptides). As shown in (Figure 3C), CD8+ T cells from DNA‒RP peptide vaccinated mice, but not CD8+ T cells from control mice, preferentially recognized B16 melanoma cells after treatment with TMZ + HSP90i.
Superior anti‒melanoma efficacy is associated with combination immunotherapy using TMZ/HSP90i and adoptive CD8+ T cells reactive against DNA‒RP
Given the ability of TMZ to enhance melanoma expression of DNA‒RP, and for the HSP90i STA9090 to promote enhanced recognition of TMZ‒treated melanoma cells by anti‒DNA‒RP CD8+ T cells in vitro, we hypothesized that TMZ + STA9090 pre‒conditioning would sensitize progressively growing B16 melanomas to the subsequent anti‒tumor action of adoptively‒transferred CD8+ T cells reactive against DNA‒RP‒derived peptide epitopes. For these experiments, B16 melanomas were established s.c. in C57BL/6 mice for 6 days, at which time they received i.p. TMZ conditioning for 2 consecutive days, followed by weekly i.p. administration of STA9090 (25 mg/kg; or the DMSO vehicle control) beginning on days 8 and 15 post‒tumor inoculation (Figure 4A). One day following treatment with the HSP90i or DMSO vehicle (i.e. days 9 and 16 post‒tumor inoculation), the mice received i.v. injection of 1 x 106 fluorescently (CFSE)‒labeled anti‒DNA‒RP CD8+ T cells (isolated from the spleens of C57BL/6 vaccinated against pooled DNA‒RP peptides, as in (Figure 4). As shown in (Figure 4B), while treatment of TMZ‒conditioned B16‒bearing mice with either STA9090 alone or anti‒DNA‒RP CD8+ T cells alone failed to impact tumor growth, the combined therapy (STA9090 + adoptive cell therapy; ACT) resulted in significantly reduced tumor progression. As expected, therapeutic intervention with TMZ/STA9090 (+/‒ ACT) led to a reduction in day 17 tumor expression of DNA‒RP (such as MRE11) in vivo (Figure 4C). Interestingly, the combined TMZ + ACT treatment protocol also resulted in less MRE11 expression in tumors (when compared to the TMZ only treatment cohort of mice (Figure 4C), possibly due to immune selection of tumor cell sub‒populations by vaccine‒induced anti‒MRE11 CD8+ T cells used for ACT). We also observed the greatest degree of cellular apoptosis within tumors of mice treated with TMZ/HSP90i + ACT (Figure 4D); p < 0.05 vs. all other cohorts, ANOVA, which was associated with the superior infiltration of tumors by CFSE+ ACT T cells (Figure 5A) and Type‒1 Tbet+CD8+ T cells (Figure 5B).
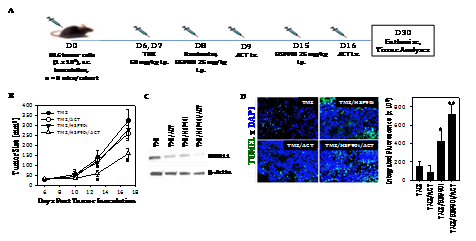
Figure 4 Combined TMZ/HSP90i + ACT therapy of established B16 melanomas is superior to chemo‒ or ACT‒monotherapy in reducing tumor growth in vivo. As depicted in the panel A schema, C57BL/6 mice were injected s.c. in their right flanks with 105 B16 melanoma cells and tumor allowed to establish for 6 days. All animals were conditioned with TMZ (50 mg/kg) for 2 days, before then randomizing them into 8 mice/cohort (i.e. each group with a comparable mean tumor size of approximately 40‒50 mm2) on day 8. Tumor‒bearing mice were then treated with HSP90i (STA9090; 25 mg/kg, i.p., on days 8 and 15) or vehicle control (DMSO) +/‒ ACT (106 splenic CD8+ T cells harvested from DNA‒RP peptide vaccinated mice as in Fig. 3A injected i.v. into the tail vein on days 9 and 16). Tumor growth was then monitored every 3‒4 days through day 30 (euthanasia) and is reported as mean + SD tumor size in mm2 (B); *p < 0.05 vs. all other cohorts (ANOVA). Animals bearing day 30 tumors of median size were subsequently harvested and analyzed for expression of DNA‒RP MRE11 by Western Blotting (C) and for cellular apoptosis by TUNEL staining as imaged and quantitated using fluorescence microscopy and Metamorph software, per Materials and Methods (D). *p < 0.05 vs. TMZ alone; **p < 0.05 vs. all other cohorts (ANOVA). All results are reflective of 3 independent experiments performed in each instance.
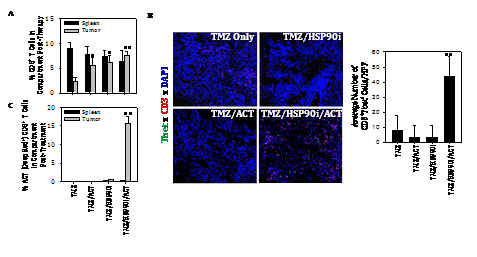
Figure 5 Combined TMZ/HSP90i + ACT therapy of established B16 melanomas promotes superior tumor infiltration by Type‒1 CD8+ TIL. Day 30 harvested tumors from (Figure 4) studies were analyzed by flow cytometry (panel A, for CD8+ TIL) or fluorescence microscopy (B, for Tbet+CD3+ TIL) as outlined in Materials and Methods. Results are reflective of 3 independent experiments performed. *p < 0.05 versus **p < 0.05 vs. all other cohorts (ANOVA).
Our major novel findings in this report are that DNA‒RP expressed by melanoma cells can be upregulated by treatment with genotoxic agents such as TMZ, and subsequently be induced to undergo degradation by HSP90i, leading to the improved recognition of tumor cells by DNA‒RP‒specific CD8+ T cells in vitro and in vivo. As a consequence, despite the known inherent resistance of human melanomas to TMZ,28‒30 which we also observed in vivo in B16 models (Figure 1B), this chemotherapeutic agent can be used to “accumulate” higher levels of DNA‒RP to serve as conditional supply of immunogenic peptides when used in combination with HSP90i such as STA9090. This tumor conditioning effect was not restricted to TMZ, as we also observed that the genotoxic anthracycline doxorubicin,31,32 but not the BRAFi dabrafenib, was also able to promote elevated expression of DNA‒RP in treated melanoma cells that were consequently susceptible to HSP90i‒induced degradation (Figure S1).
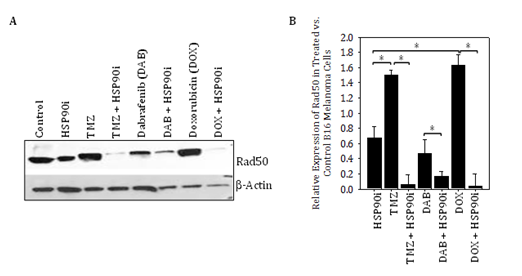
Figure S1 Genotoxic chemotherapy agents, but not BRAFi dabrafenib, promote increased expression of DNA‒RP Rad50 in B16 melanoma cells that is induced to undergo degradation by HSP90i in vitro. In A, the B16 murine melanoma cell line was incubated in the absence or presence of TMZ (50 μM), dabrafenib (20 μM) or doxorubicin (0.1 μM) +/‒ STA9090 (25 μM) for 12h at 37oC, prior to cell solubilization and Western Blotting analysis as outlined in Figure 1 for the DNA‒RP Rad50 versus b‒actin control proteins. In B, relative expression of Rad50 in treated vs. control (untreated) B16 melanoma cells is plotted based on data from Figure S1A. Results are reflective of 3 independent experiments performed. *p < 0.05 for the indicated differences between groups (t‒test).
The finding that we could induce CD8+ T cell responses against non‒mutated “self” peptides derived from DNA‒RP using DC‒based vaccines is consistent with previous reports supporting the ability of melanoma lineage antigen (MART1, gp100, tyrosinase, TRP1/2 and others)‒based vaccines to activate a low‒to‒moderate avidity T cell repertoire in mice and humans, despite the host’s apparent operational “self‒tolerance” to such antigens.27,33‒35 Although we only analyzed the capacity of combination TMZ/HSP90i‒based therapy to improve the anti‒tumor efficacy of adoptively‒transferred anti‒DNA‒RP CD8+ T cells in our melanoma treatment models, we would anticipate that this drug combination would also improve the therapeutic benefits associated with active specific vaccination against DNA‒RP (using antigen‒loaded Type‒1‒polarized DC or alternate strategies).
Such analyses will be pursued in future studies extended to include alternate genotoxic agents (including DOX known to promote tumor “immunogenic cell death Obeid et al.36 and additional inhibitors of HSP90 (including alternate in‒clinic HSP90i that affect ATP‒binding Butler et al.37 as well as, HDACi known to alter the acetylation of regulatory lysine residues in the HSP90 protein leading to its dysfunction.38‒40 Although we failed to observe any evidence for pathologic autoimmunity in our ACT modeling, it will be important to monitor mice vaccinated against DNA‒RP in future experiments for deleterious autoimmune sequelae, particularly in combination approaches designed to promote strong inflammatory responses.3‒5,41‒43
Our findings also support the ability of combined TMZ/HSP90i + ACT‒based therapy to promote enhanced infiltration of melanomas by Type‒1 (Tbet+) CD8+ T cells (including the ACT population of CSFE‒labeled CD8+ T cells) in association with improved treatment outcome, and are consistent with several recent reports supporting higher numbers of CD8+ TIL as a predictive biomarker for patient benefit to immunotherapy.44‒47 However, as these CD8+ T effector TIL produce IFN‒g (based on ELISA results), which can promote upregulated expression of immune checkpoint molecules such as PD‒L1 Tokito et al.46 and Mandai et al.48 in the TME. PD‒L1 agonism of PD‒1 on TIL can reduce the viability and anti‒tumor functionality of these protective T cell populations in vivo.4, 5 Our preliminary data suggest that PD‒L1 expression is indeed upregulated preferentially in the TME of mice treated with combined TMZ/HSP90i + ACT (Figure S2). Hence, future studies will be designed to determine whether the anti‒PD‒L1 and/or anti‒PD‒1 immune check point inhibitor antibodies can further improve the anti‒tumor efficacy of our current combination chemoimmunotherapy approach.
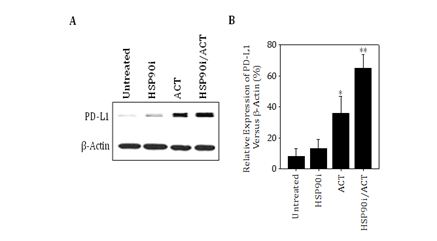
Figure S2 B16 melanomas treated with combined TMZ/HSP90i + ACT therapy express higher levels of PD‒L1 in vivo. Day 30 harvested tumors from Figure 4 studies were analyzed by immunofluorescence microscopy for expression of PD‒L1 as outlined in Materials and Methods. Results are reflective of 3 independent experiments performed. *p < 0.05 vs. untreatedcontrol tumors; **p < 0.05 vs. all other cohorts (ANOVA).
The ability of HSP90i to enhance recognition of melanoma target cells by anti‒DNA‒RP CD8+ T cells might yet be suboptimal, since cellular expression of the HSP70 chaperone molecule is known to be increased in response to HSP90i Whitesell et al.49 and HSP70 can substitute for HSP90 in stabilizing client proteins.50 Such complementation in the molecular salvage pathway might limit the pool of DNA‒RP accessible for processing, and hence, the amount of DNA‒RP‒derived peptides capable of being presented to anti‒DNA‒RP CD8+ T cells. As a consequence, it might be anticipated that transient coordinate application of HSP70i + HSP90i might result in more robust anti‒tumor activity mediated by DNA‒RP‒specific T effector cells, as well as, to a greater degree of tumor cell apoptosis. Given concerns for systemic toxicities that might arise in such combination regimens, these treatment protocols will need to be carefully evaluated in future studies.
In conclusion, we believe that our translational results may support the design of new chemoimmunotherapeutic options for patients with melanoma or alternate advanced‒stage forms of solid cancer that exhibit chemotherapy‒resistance and/or that fail current first‒line therapies (including immune checkpoint blockade).
The authors wish to thank Drs. Susanne Gollin, Robert Binder and Quasar Padiath for their careful review and suggestions made during the preparation of this manuscript. This project used the UPCI’s Peptide Synthesis, Protein Sequencing and Vector Core Facilities that are supported in part by NIH P30 CA047904. The performance of this work was also supported by NIH R01 CA169118 (to W.J.S.).
The authors declare that there are no financial interests or conflicts of interest.

©2017 Storkus, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.