MOJ
eISSN: 2373-4442


Short Communication Volume 4 Issue 1
1Department of Microbiology and Immunology, Center for Molecular Virology and Translational Neuroscience, Institute for Molecular Medicine and Infectious Disease, Drexel University College of Medicine, Philadelphia, PA, USA
2Department of Medicine, Division of Infectious Diseases and HIV Medicine, Center for Clinical and Translational Research, Institute for Molecular Medicine and Infectious Disease, Drexel University College of Medicine, Philadelphia, PA, USA
3Sidney Kimmel Cancer Center, Thomas Jefferson University, Philadelphia, PA, USA
Correspondence: Brian Wigdahl, Professor and Chair Department of Microbiology and Immunology Director, Institute for Molecular Medicine and Infectious Disease, Drexel University College of Medicine, 245 N. 15th Street, MS # 1013A, Philadelphia, PA 19102, USA, Tel 215 762-7598, Fax (215) 762-1955
Received: September 15, 2016 | Published: September 19, 2016
Citation: Parikh N, Dampier W, Passic SR, Zhong W, Blakey B, et al. (2016) Coinfection with Hepatitis C Virus among HIV-1-Infected Individuals Increases Proinflammatory Cytokines and HIV-1 Disease Severity. MOJ Immunol 4(1): 00115. DOI: 10.15406/moji.2016.04.00115
HIV-1, along with hepatitis B and C viruses (HBV and HCV), are known to share similar routes of transmission, including intravenous drug use, blood transfusions, sexual intercourse, and perinatal exposure. Thus, coinfection with HIV-1 and HBV or HCV is common. HIV-1 impacts the natural course of HBV and HCV infection by accelerating the progression of HBV/HCV-associated liver disease toward end-stage cirrhosis and quantitative depletion of the CD4+ T-cell compartment. HBV or HCV coinfection with HIV-1 has also been associated with increased mortality when compared to either infection alone. In particular, we focus on the impact of coinfection with HCV on the HIV-1 pathogenic process in patients in the Drexel Medicine CNS AIDS Research and Eradication Study (CARES) Cohort. Comparing HIV-1 mono-infection with HIV-1/HCV coinfection allows defining the detrimental effect of HCV coinfection as it relates to viral pathogenesis, disease progression, and the associated immune response during the course of this complex interplay, in the background of drug abuse, which is an important risk factor to both HIV-1 and HCV infection and on HIV-1 disease severity. The uniqueness of this study includes the fact that the patients are part of a cohort of HIV-1-infected individuals whose drug histories have been recorded longitudinally.
Keywords: HIV, HCV, Cytokines, Coinfection, HIV-1, HCV coinfection
HBV, hepatitis B virus; HCV, hepatitis C virus; NK, natural killer; DCs, dendritic cells; HIV-1, human immunodeficiency virus type 1; CNS, central nervous system; ART, anti-retroviral therapy; LME, linear mixed effects
It is essential to understand the natural course of hepatitis C virus (HCV) infection. HCV is a single-stranded RNA virus belonging to the Flaviviridae family.1 It has been shown to be a major cause of chronic hepatic fibrosis that can progress to cirrhosis and hepatocellular carcinoma.2 Once infected with HCV, about 70% of the infected individuals are not able to clear the infection and thus the virus persists in the body leading to chronic infection. The remaining 30% of infections involve acute infection that is spontaneously cleared.3
However, in cases of HIV-1 and HCV coinfection, the number of chronic HCV infections rises to 90%.4 The result of any viral infection depends on the interplay between the activation of host cellular factors to the infection and the viral mechanisms that counteract them.5 HCV is responsible for activating antiviral interferon (IFN)-α/β expression, but irrespective of the expression of these cytokines, HCV can still replicate within the liver.6 This could be due to the fact that the nonstructural proteins, nonstructural protein (NS)3 and NS5A, and the structural protein E2 can block the expression as well as transcription of IFN-α/β genes. Also, the NS5A protein induces expression of interleukin (IL)-8, associated with IFN-α inhibition7 HCV can also block the antiviral action of natural killer (NK) and NKT cells thus hindering the expression of IFN-γ (an antiviral cytokine) due to the interaction between E2 and NK-cell CD81 molecule.8 The production of cytokines by dendritic cells (DCs) is particularly affected by chronic HCV infection and there are contradictory reports concerning the expression of Th1 cytokines, with some studies suggesting that a Th1 response is suppressed9 and some studies suggesting a progressive liver injury during chronic HCV has been associated with an increase in the Th1 response-associated cytokines.10 In this regard, HCV CD4+ T cells also play an important role in providing an adaptive response by activating cytotoxic and humoral responses. This can involve the secretion of IFN-γ, which can recruit neutrophils and macrophages, contributing to an inflammatory response. However, these CD4+ T cells may additionally release Th2 cytokines such as IL-4 and IL-10, thus, limiting the Th1 response.11 It is observed that while; a specific Th1 response has been observed in HCV infections,12 within a chronic HCV infection there is a definite shift to a Th2 response, contributing to an imbalance in the Th1/Th2 response profile.5,13
HIV-1 and HCV share similar modes of transmission. Comparable to transmission of human immunodeficiency virus type 1 (HIV-1), HCV can be transmitted via sexual contact as well as by mother to child vertical transmission. In addition, intravenous drug use (IVDU) also plays a significant role in transmission based on sharing of contaminated needles, and other injection paraphernalia1 as well as risky sexual behavior.14 Globally, based on the 35 million people infected with HIV-1, it is estimated that approximately 7 million people suffer from chronic HCV infection, which is about 20% of the HIV-1-infected population.15 Alarmingly, HIV-1-infected individuals are at an increased risk of dying due to coinfection with chronic HCV in addition to other acquired immunodeficiency syndrome (AIDS)-related complications. In areas of the world where highly active antiretroviral therapy (HAART) is available, following AIDS-related complications, chronic infection with HCV has now become the leading cause of death among HIV-1-infected individuals.16 HIV-1 is known to lead to a more aggressive form of HCV disease, increasing the frequency of cirrhosis a causative factor in HCV-related mortality.14,17 While a detailed molecular mechanism is unknown concerning the precise ways in which HIV-1 affects HCV pathogenesis, it is known that reduced CD4+ and CD8+ T-cell responses18 and microbial translocation can advance hepatic fibrosis.19
In addition, the varied immune response during HIV-1 and HCV infections as well as modulation of certain cytokines and chemokines such as IFN-γ, MCP-3, and MIP-3α, are significant factors in the altered pathogenesis of HIV-1 and HCV coinfection.20,21 In a decade long study comparing HIV-1 mono-infected individuals versus HIV-1 and HCV co-infected individuals, the co-infected individuals showed an increased prevalence of multiple neurologic disorders, 60% in the co-infected individuals as compared to the 46% in the HIV-1 mono-infected individuals. The neurocognitive disorders were also found to be more severe within the co-infected individuals.22 This study has been supported by the fact that HCV has been detected in the central nervous system (CNS),23 cerebrospinal fluid24 and peripheral nerves of HCV-infected individuals.25 This study highlighted the adverse effects of HCV as comorbid factor to HIV-1 disease with respect to neurocognitive impairment. Thus, it is necessary to comprehend the factors that are essential to HIV-1 and HCV coinfection pathogenesis, identify molecular markers to identify progression of disease and accordingly design therapeutic strategies that aim at lowering the morbidity associated with this coinfection.
It is crucial to treat chronic HCV in HIV-1 and HCV coinfected individuals, due to the rapid progression to end-stage liver disease within these individuals15,26 and reduced tolerance of antiretroviral agents which is associated with greater hepatotoxicity.1,27 It has been shown that treatment of chronic HCV can lead to reduced hepatotoxicity28 and regression of the drawbacks such as liver fibrosis.15,29,30 While HCV genotype and viral load can influence the response to HCV therapy, extent of hepatic fibrosis and CD4 counts are most indicative of the individuals that should be receiving HCV therapy.15 HCV therapy is usually initiated in individuals who have more than 200-350 CD4+ T cells/mm3. However, in individuals who are on HAART and with CD4+ T-cell counts below 200 cells/mm3, treating HCV is a challenging task, since there are other factors such as, severity of liver disease, extent of HIV suppression and time since being termed as being HCV-positive, that have to be taken into consideration.15,31 Thus, it is evident that within co-infected individuals, treatment options are varied as compared to individuals with HCV mono-infection.
At the time this data was analyzed, 504 HIV-1-infected patients were enrolled in the Drexel Medicine CARES Cohort that were being cared for in the Drexel University College of Medicine HIV/AIDS Partnership Care Practice, which is located in Philadelphia, Pennsylvania, USA. Utilizing this patient population, the goal of this study was to understand the impact of HIV-1/HCV coinfection on HIV-1 disease progression within these individuals. To achieve this, a detailed assessment of their drug, alcohol and tobacco history and current use was assessed, followed by collection of whole blood from these patients, which was then used for plasma and PBMC isolation. Plasma samples were used to perform Luminex based-cytokine profiling assays to understand the impact of HIV-1/HCV coinfection in a population of preferential drug users as compared to a population of preferential non-drug users (PN). A population of multidrug users (MDU) using cocaine, cannabinoid and benzodiazepines were also included in the study to understand the impact of coinfection while abusing a diverse range of illicit drugs. Further analyses were done to correlate the cytokine profiles to various clinical parameters such as viral load, CD4+ and CD8+ T-cell count and CD4: CD8 ratio.
Patient recruitment, clinical data and sample collection
Patients in the Drexel Medicine CARES Cohort have been recruited from the Partnership Comprehensive Care Practice of the Division of Infectious Disease and HIV Medicine at Drexel University College of Medicine, which is located in Center City Philadelphia, Pennsylvania, USA as previously described.32 The study was approved by the Drexel University College of Medicine Institutional Review Board under protocol 1201000748 (Brian Wigdahl, principal investigator) in compliance with Helsinki Declaration of 1975 as revised in 2000. All participants were required to sign an Informed Consent Form. Once enrolled, an anonymous identifier was assigned and their clinical data was collected and entered onto individual Case Report Forms. Clinical information collected directly from the patient included age, gender, year of birth, ethnicity, race, HIV exposure category, history of illicit drug use, tobacco and alcohol use, potential pregnancy, and approximate year of seroconversion. Other additional information was gathered from the patient’s clinical history chart, such as initial, nadir, and current CD4+ T-cell counts; initial, nadir and current CD8+ T-cell counts; peak and current viral loads; disease status; current anti-retroviral therapy (ART); hepatitis B and hepatitis C virus coinfection; AIDS-defining illnesses; notations on mental health history including inpatient stays for mental health reasons; history of depression, schizophrenia, other neurological diseases; and any other notations on other complicating conditions related to HIV-1 infection. Blood samples were drawn from each patient at initial and each subsequent return visits. Blood samples were used for drug screening, plasma analysis, and PBMC isolation. Each patient was called approximately every six months for a return visit to allow for a longitudinal study with at least one recall per year. Each patient was tested for presence of various drugs at every longitudinal visit, thus, allowing for detailed histories of drug abuse recorded. Patients were classified into drug-user groups based solely on drug testing results; their verbal admissions or denials were not considered.
Patient drug screening
Blood samples from each patient’s first and every subsequent visit were screened for a standard seven-drug profile drug screen (LabCorp, Burlington, NC), which includes amphetamines (amphetamine and metamphetamine); barbiturates (amobarbital, butalbital, pentobarbital, phenobarbital, and secobarbital); benzodiazepines (desalkylflurazepam, flurazepam, and diazepam); cannabinoids (tetrahydrocannabinol and tetrahydrocannabinolic acid); cocaine (cocaine and benzoylecgonine); opiates (codeine and morphine); and phenylcyclidine.
PBMC isolation and plasma sample storage
Three purple-top blood collection tubes (Vacutainer; Becton, Dickinson, Franklin Lakes, NJ) containing EDTA were used to collect 50 ml of whole blood from patients for plasma and PBMC isolation. Samples underwent an initial centrifugation for plasma isolation. Approximately 9 mL of plasma was collected per patient and 1mL aliquots were made, which were placed at -85°C for long-term storage and were used as needed. Patient-derived PBMCs were then isolated from whole blood using Ficoll-hypaque (Amersham Biosciences, Piscataway, NJ, USA) density gradient centrifugation as previously described (Pirrone et al., 2010). Genomic DNA and total RNA was isolated using a Qiagen procedure according to manufacturer guidelines (Qiagen, Valencia, CA, USA)
Luminex 30 plex assay
The Human Cytokine 30-plex panel for the Luminex platform (Life Technologies, Grand Island, NY) was used to quantify the indicated cytokines, chemokines and growth factors present in the plasma samples obtained from the patients within the Drexel Medicine CARES Cohort. The multiplex immunoassay was performed as described by the manufacturer and the plate was read on the Luminex 200 system (Luminex Corporation, Austin, Texas). Briefly, the plasma samples were thawed on ice and clarified by centrifugation (1000 x g for 10 minutes) before they were used in the assay so as to prevent clogging of the filter plate. The plasma sample (50μl) was diluted with 50 μl of assay diluent to ensure a 1:2 dilution as described by the manufacturer (Life Technologies, Grand Island, NY). The wells were pre-wet with wash solution (200μl), incubated for 30 sec, aspirated and blotted on paper towels. Bead solution (25μl) was then added to each well, followed by wash solution (200μl). The beads were allowed to soak for 30 sec under foil, away from the light. This was followed by aspiration and blotting on the paper towels. Incubation buffer (50μl) was added to each well and to the standard wells; standard was added (100μl), while the sample wells received assay diluent (50μl) and diluted plasma sample (50μl). The plate was covered and allowed to shake for 2 hr. The plate was aspirated, washed twice and a biotinylated antibody (100μl) was added to all the wells, following which the plate was covered and incubated for 1 hr on the shaker. The plate was aspirated, washed twice and streptavidin-bound RPE (100μl) was added to each well, followed by incubation for 30 min on the shaker. The plate was then aspirated, washed three times and dried. Wash solution (100μl) was added to each well, shaken for 3 min and read on the Luminex 200 system (Luminex Corporation, Austin, Texas). Each multiplex immunoassay was performed twice in duplicate, giving 4 values for each cytokine concentration.
Biostatistical analysis
To begin the experimental analysis, all Luminex values that fell outside the standard curve were discarded. Subsequently, all remaining raw cytokine values were quantile transformed for each array using the normalize.quantiles function in the Bioconductor package.33 A linear mixed effects (LME) model was used as one analysis and included terms for age, gender, and HAART status. The dosage response of drugs was included in a second analysis using the weighted linear contribution model (WLCM). This model was based on the linear mixed-effects model and included terms for age, gender, HAART status, and drug abuse as confounding variables. Age was considered as a linear variable whereas gender, HAART status (continuous, discontinuous, and naïve), and drug abuse as treated as a linear variable as described below.
In LME, the HIV-1-monoinfected patients and HIV-1/HCV co-infected were compared with each other in the absence of drug abuse. The algorithm for the LME model is as follows: Cytokine = ĸ (age) + ĸ (gender) + ĸ (HAART) + ĸ (HCV).
The WLCM attempts to model the effect of drug use on the dependent variable by considering each drug as a linear contributor, while still comparing the HIV-1-monoinfected patients and HIV-1/HCV co-infected patients. Results from all patients with results from the cytokine analysis were used to build the WLCM model, regardless of their drug use. This approach provided a methodology to analyze patients with varying levels of drug usage. The inclusion of patients using multiple drugs allows the algorithm to estimate the effect of several drugs within a multidrug use scenario and its possible impact on mono-infected versus co-infected patients. The algorithm for the WLCM model is as follows: Cytokine = ĸ (age) + ĸ (gender) + ĸ (HAART) + ĸ (HCV) + ĸ (cocaine) + ĸ (cannabinoid) + ĸ (benzodiazepine). In the WLCM model drug use was defined as the fraction of times the patient tested positive over all visits to date as previously described.34
Statistical analysis was performed using R2.15.1 (The R Foundation for Statistical Computing) and a p-value less than 0.05 was considered significant.
HIV-1/HCV coinfected subcohort demographics and clinical parameters
Previously from the 504 HIV-1-infected individuals in the Drexel Medicine CARES Cohort, we selected a subcohort of 102 patients based on longitudinal drug use and race.34 This subcohort of patients was used here to define a HIV-1/HCV co-infected subcohort. Patients were classified as HIV-1 mono-infected only if they tested positive for HIV-1 and not for HCV. Patients were classified as HIV-1/HCV co-infected only if they tested positive for HIV-1 and HCV. HCV infection was determined using the latest clinical history of every patient. A number of patients were also tested independently using their plasma for HCV viral load using Real Time PCR (LabCorp, Burlington, NC). The results from these tests correlated with HCV infection status retrieved from the clinical histories of the patients (data not shown). There were 58 patients included in the mono-infected population and 44 patients included in the co-infected population for a total of 102 patients. One patient could not be appropriately classified. About 59% of the HIV-1/HCV coinfection subcohort was male, while the remaining 41% were female. Greater than 84% patients were on continuous HAART (cH), almost 5% of patients were discontinuous with respect to HAART (dH) and 10.5% of patients were naïve to HAART (nH). The average age of the subcohort was 47 years and the number of years since they were diagnosed as being seropositive was an average of 11.6 years (Table 1).
Demographic variables |
Categories |
Black/AA count (%)/Mean (±SD) with clinical variables |
Gender |
Male |
60 (58.8%) |
Female |
42 (41.2 %) |
|
Drug Use |
Tobacco |
60 (58.8%) |
Alcohol |
45 (42.2%) |
|
Amphetamines |
0 |
|
Barbiturates |
0 |
|
Benzodiazepines |
6 (5.9%) |
|
Cannabinoids |
18 (17.6%) |
|
Cocaine+Metabolite |
44 (43.1%) |
|
Opiates |
0 |
|
Phenylcyclidine |
0 |
|
HAART Status |
cH |
86 (84.3%) |
dH |
5 (4.9%) |
|
nH |
11 (10.8%) |
|
Age |
47 (±9.5) |
|
Years since Diagnosis |
11.6 (±6.8) |
|
HCV Status |
Positive |
44 (43.1%) |
Negative |
58 (56.9%) |
Table 1 HIV-1/HCV coinfection subcohort demographics. The demographics of HIV-1/HCV coinfection subcohort are shown. From a total of 102 black/AA patients, 58 patients were HIV-1 mono-infected and 44 patients were HIV-1/HCV co-infected. Sixty patients were male and 42 were female. The alcohol, tobacco and drug use history of the subcohort was documented. Eighty six patients were on continuous HAART (cH), 5 patients were on discontinuous HAART (dH) and 11 patients were naïve to HAART (nH). The average age of the subcohort was 47 years and 11.6 years was the average age since they were diagnosed positive for HIV-1 infection
HAART, highly active antiretroviral therapy; SD, standard deviation; cH, currently on HAART; dH, discontinuous HAART; nH, Naïve to HAART.
Within these patients, the mean CD4+ T-cell count of the mono-infected patients was 538 cells/μl, which was greater than the mean CD4+ T-cell count of the co-infected patients, which was 484 cells/μl reflecting the fact that the mono-infected patients are relatively healthier compared to the co-infected patients. The latest mean CD8+ T-cell count of the mono-infected patients was lower at 1,028 cells/μl as compared to the mean CD8+ T-cell count of the co-infected patients, which was 1,174cells/μl. This was reflected in the CD4:CD8 ratios, wherein mono-infected individuals had higher mean ratios (0.635) than the co-infected individuals (0.541). The viral load of the subcohort was log transformed and plotted and showed that the mono-infected individuals had lower mean viral loads (2.423) than the co-infected individuals (Figure 1). A normal CD4+ T-cell count ranges from 500 - 1,000 cells/μl while a normal CD8+ T-cell count ranges from 300 - 1,000 cells/μl.35 Investigators from the US Military’s HIV Natural History study have reported that elevated CD8+ T-cell count while a patient is on HAART could be a warning for treatment failure and has been associated with accelerated HIV disease progression.36 Thus, it seems that overall, the mono-infected patients were healthier as compared to the co-infected patients, especially while considering the decreased CD4+ T-cell count, increased CD8+ T-cell count and viral loads, underlining the negative impact that HCV coinfection has on HIV pathogenesis.
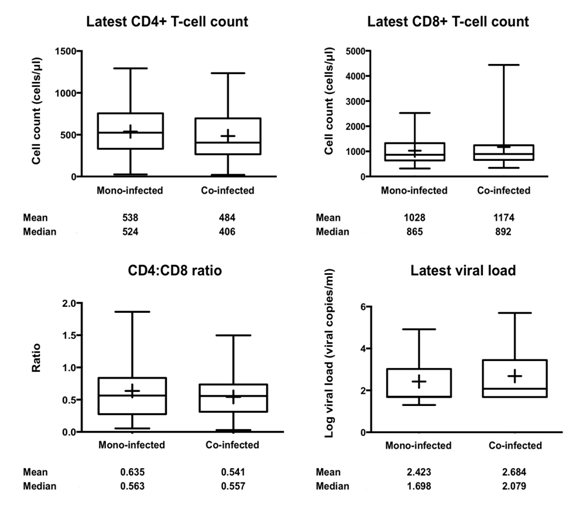
Figure 1 Clinical parameters for the HIV-1/HCV coinfection subcohort. Using Box-Whisker plots, the latest CD4+ and CD8+ T-cell counts, CD4:CD8 ratios, and the latest viral load of the coinfection subcohort are shown which reflect the range and health of the subcohort. The mean (+) and median values for each are provided. A standard student’s t-test was performed. The latest viral loads were log10 transformed for generating plots. A p value less than 0.05 was considered statistically significant.
Impact of HCV coinfection on cytokine profiles in patients infected with HIV-1
To understand the impact of HCV coinfection on cytokine profiles in patients within the HIV-1/HCV co-infected subcohort, the plasma samples from the mono-infected and co-infected patients were subjected to a human cytokine 30-plex panel immunoassay. The 30 cytokines were then examined with respect to mono-infection of HIV-1 or HIV-1/HCV coinfection utilizing 2 biostatistical models, LME (n=102) and WLCM (n=102). The LME model takes into account the effect of standard confounders such as age, gender and HAART status, while the WLCM takes into account the effect of the same standard confounders as well as the entire history of drug abuse of the patients. The Cytokine Human 30-plex panel used on the Luminex 200 platform was specifically designed to quantify human cytokines, chemokines and growth factors in serum, plasma and tissue culture cell supernatant. It uses the xMAP technology (multi-analyte profiling), which is used for multiplex format.
Analysis using the LME model identified eotaxin (p=0.0358), interferon-gamma (IFN-γ) (p=0.0088), interleukin-17 (IL-17) (p=0.0255), interleukin-4 (IL-4) (p=0.0218), interferon gamma-induced protein 10 (IP-10), also known as CXCL10, (p=0.0002), monocyte chemotactic protein-1 (MCP-1) (p=0.0482), tumor necrosis factor-alpha (TNF-α) (p=0.0007) and the collective Th1 cytokine panel, values of three cytokines, IFN-γ, IL-2 and TNF-α, added together, as being significantly associated with HCV coinfection, while granulocyte colony stimulating factor (G-CSF) (p=0.0654), granulocyte-macrophage colony stimulating factor (GM-CSF) (p=0.0666), interleukin-10 (IL-10) (p=0.0663) and macrophage inflammatory protein-1 beta (MIP-1β) (p=0.0962) trended to be associated with HCV coinfection (Figure 2). From the cytokines that had significant p-values, IP-10, was the only cytokine that was positively associated with HCV coinfection, which has been shown in previous studies to be upregulated during HCV infection,37,38 while all the seven other cytokines were significantly negatively associated with HCV coinfection. From the cytokines that trended to be associated, G-CSF and MIP-1β were negatively associated with HCV coinfection and GM-CSF and IL-10 were positively associated (Figure 2).
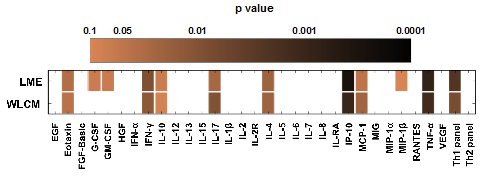
Figure 2 Cytokines significantly associated with HCV coinfection. Cytokines analyzed using the linear mixed effects model (LME) (n=102) adjusting for age, gender and highly active antiretroviral therapy (HAART) status as confounders; and the weighted linear contribution model (WLCM) (n=102), adjusting for age, gender, highly active antiretroviral therapy (HAART) status, and drug use as confounders, are expressed as a heat map with respect to their p-values. The p values are denoted in log scale and as the color gets darker, the p value becomes more significant. Values of p < 0.05 were considered statistically significant. Cytokines with a p value less than 0.1 are shown. Cells that lack color had p values greater than 0.1. The Th1 panel consists of interferon-γ (IFN-γ), interleukin-2 (IL-2), and tumor necrosis factor-α (TNF-α), while the Th2 panel consists of IL-4, IL-5, and IL-10. The results take into consideration drug use at all of the patient’s longitudinal visits. EGF, epidermal growth factor; FGF, fibroblast growth factor, HGF, human growth factor; RANTES, regulated on activation, normal T-cell expressed and secreted; and VEGF, vascular endothelial growth factor.
Analysis using the WLCM model showed similar cytokines being significantly associated with HCV coinfection. Cytokines identified were eotaxin (p=0.0436), IFN-γ (p=0.0136), IL-17 (p=0.0098), IL-4 (p=0.022), IP-10 (p=0.0009), MCP-1 (p=0.0233), TNF-α (p=0.0013) and the collective Th1 cytokine panel (p=0.0075), values of three cytokines, IFN-γ, IL-2 and TNF-α, added together, were significantly associated with HCV coinfection, while IL-10 (p=0.0859) trended to be associated with HCV coinfection. From the cytokines that had significant p-values, IP-10, was the only cytokine that was positively associated with HCV coinfection while all the other cytokines were significantly negatively associated with HCV coinfection. IL-10 trended to be positively associated with HCV coinfection (Figure 2) (Table 2).
Cytokine |
Effect (p-Value) |
Eotaxin |
-27.19 (0.0436) |
IFN-g |
-7.18 (0.0136) |
IL-10 |
19.03 (0.0859) |
IL-17 |
-2.76 (0.0098) |
IL-4 |
-11.36 (0.022) |
IP-10 |
57.29 (0.0009) |
MCP-1 |
-500.4 (0.0233) |
TNF-a |
-4.62 (0.0013) |
Th1 panel |
-13.72 (0.0075) |
Table 2 Effect sizes for all cytokines significantly associated with HIV-1/HCV coinfection using the WLCM biostatistical model. The effect signifies the size of the effect that HCV coinfection has on the value of each cytokine. A minus sign in front of the effect value denotes that the drug has a negative effect on the value of the cytokine (downregulation), while no sign in front of the effect value denotes that the drug has a positive effect on the value of the cytokine (upregulation). EGF, epidermal growth factor; GM-CSF, granulocyte-macrophage colony-stimulating factor; IFN, interferon; IL, interleukin; IP-10, interferon gamma-induced protein 10; MCP-1, monocyte chemotactic protein-1; TNF-α, tumor necrosis factor-alpha; WLCM, weighted linear contribution model
Sensitivity analysis
In order to assess the effects that the number of patient visits would potentially have on the WLCM model, a sensitivity analysis was performed. This analysis repeats the analysis performed using the WLCM model in the previous figures (Figure 2). But the analysis is now limited to patients who have been in the study for certain number of visits as indicated in the figure, since all the patients in the subcohort did not have equal number of return visits due to the ongoing longitudinal nature of the study. The results show that for all of the cytokines that were significantly associated with HCV status, the number of visits by a patient did not affect the results in most cases, thus, providing evidence that the model was accurate regardless of the number of visits per patient (Figure 3).

Figure 3 Sensitivity analysis. In this analysis, the patient set was restricted by increasing the number of visits from 1 to 3 in incremental steps of 1. The WLCM analysis was re-calculated as described previously in the Materials and Methods. This analysis excludes Eotaxin, G-CSF, and IL-10 as they are only significant at particular inclusion cutoffs.
Since, HIV-1 and HCV, share transmission risk factors, it is common for an HIV-1-infected individual to be co-infected with HCV. Within the United States, from the approximate 1.2 million HIV-1-infected individuals, about 25% of these are co-infected with HCV.39 The interplay between both the viruses is complex and usually leads to accelerated forms of HIV-1 disease as well as HCV disease.21 In fact, some recent reports have suggested that HCV-related deaths are greater than HIV-1-related deaths in the United States, and that the HIV-1/HCV co-infected individuals are the ones bearing a significant portion of the mortality.40 Accelerated liver disease, higher viral loads, and differential therapeutic regimens are some of the specific challenges encountered during HIV-1/HCV coinfection. This study explored the effects of HIV-1/HCV coinfection versus HIV-1 mono-infection on immune parameters within HIV-1-infected individuals of the Drexel Medicine CARES Cohort.
The robust WLCM allows understanding of the impact of HCV coinfection within HIV-1-infected individuals while taking into account their extensive drug abuse histories. The analysis identified that IFN-γ and IP-10 were upregulated in HIV-1/HCV co-infected individuals; while eotaxin, IL-17, IL-4, MCP-1, TNF-α and the collective Th1 panel were identified as being significantly down regulated in individuals who were HIV-1/HCV co-infected as compared with HIV-1 mono-infected individuals. Inflammatory cytokines and chemokines, are essential due to their important roles during inflammation during viral infections such as HCV. Interestingly, there are few studies that have tried to understand differences in cytokine expression levels for HIV-1 versus HIV-1/HCV in co-infected cohorts and even these examined only a few cytokines and not an extensive panel as in the current study.41,42 An Egyptian cohort study exposed PBMCs procured from HCV-seronegative or -seropositive individuals, to HCV peptides such as NS4 and NS5 and found that IFN-γ levels while increased in both the populations, the HCV-seropositive individuals had comparatively higher levels of the cytokine.43 IFN-γ is an important cytokine for innate and adaptive immunity against viral and bacterial infections.44 Cytokine levels are in part responsible for viral clearance. And high levels of this particular cytokine in the CARES Cohort, may relate to immune responses trying to control HCV viremia.43 Another cohort study has shown that IFN-γ levels were higher in HIV-1 mono-infected individuals as compared to HCV mono-infection or HIV-1/HCV coinfection.41 However, these studies were performed with a very limited number of patients and also, no consideration was given to the drug status of these patients, which is an important confounding effect to consider when analyzing such results. Meanwhile, IP-10 has been a widely studied cytokine with respect to HCV infection. As the name suggests, it is a chemokine, inducible by IFN-γ45 and produced by several cell types such as hepatocytes, NK cells, monocytes, and T-lymphocytes.46 During HCV infection, hepatocytes produce IP-10 and circulating levels of IP-10 are known to be measurable during chronic HCV infection.37 High levels of IP-10 levels are known to be associated with reduced rates of sustained virological response during treatment of HCV with pegylated (PEG)-IFN/ribavirin (RBV)37,47–49 HIV-1/HCV coinfection38,50 and reduced spontaneous clearance to acute infection.51 Further, high IP-10 levels have been associated with greater inflammation and liver fibrosis48,49 even in a cohort of African-American injection drug users with chronic HCV.52 Thus, these increased levels of IP-10 within the HIV-1/HCV co-infected individuals in the CARES Cohort could likely be a marker of reduced response to therapy, immunologic dysfunction ultimately leading to an accelerated progression of disease.
Of the other cytokines that were negatively associated with HCV coinfection, pro-inflammatory cytokines such as TNF-α and IL-17 were included. IL-17 and TNF-α can act synergistically with each other and are involved in systemic inflammation53–55 IL-17 can recruit monocytes, neutrophils and T-helper cells to the site of inflammation56 and TNF-α is predominantly produced by macrophages, CD4+ lymphocytes, NK cells, and neurons.57 TNF-α was found to be present in increased levels when human hepatic cells were infected with HCV and HIV-1 in vitro, and this effect was further exacerbated in the presence of morphine.58 However, we have observed a decrease in the cytokine in the presence of a coinfection scenario within a drug abusing population, albeit cocaine and cannabinoid use. At the same time, IL-4, an anti-inflammatory, Th2 cytokine, has been shown to be responsible for differentiation of Th0 cells to Th2 cells.59,60 It can inhibit differentiation into Th1 cells and also Th1 cytokines and is a key regulator of humoral and adaptive immunity.61 Also, the collective Th1 panel, IFN-γ, IL-2, TNF-α, was down regulated in HIV-1/HCV co-infected individuals. There is not enough evidence or there is contradictory evidence of the levels of these cytokines within either HIV-1 or HCV mono-infection and HIV-1/HCV coinfection. However collectively, all studies point to a global dysregulation of several cytokines as well as disruption of balance of pro- versus anti-inflammatory and Th1 versus Th2 cytokines.
In summary, few studies have examined the effects of HIV-1 and HCV, together, on cytokine production and how these could affect disease progression, and effects of therapies for both the infections. It is a complex interaction that occurs between the two viruses and leads to a distortion of the delicate balance and regulation of a number of cytokines. An important future direction would be to define the regulation of cytokines with respect to aviremic versus viremic individuals in the existing cohort. Understanding how these viruses influence cytokines and their effect on the pathobiology of the disease is important and would able us to treat both the infections in a coinfection model especially within a drug using population.
None.
The authors declare there is no conflict of interests.
None.

©2016 Parikh, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.