MOJ
eISSN: 2373-4442


Research Article Volume 3 Issue 1
1Assistant Prof, PhD in Medical virology, Department of medical Microbiology, Kerman University of medical sciences, IRAN
2Department of medical Microbiology, Kerman University of medical sciences, Iran
3Assistant Prof, Department of Arboviruses & Viral Hemorrhagic Fevers, Pasteur Institute of Iran (IPI), Iran
Correspondence: Hamid Reza Mollaei, Assistant Prof, PhD in Medical virology, Department of medical Microbiology, Kerman university of medical sciences, Kerman, Iran, Tel 98-34-3257542, Fax 98-34-3257542
Received: December 26, 2015 | Published: January 4, 2016
Citation: Mollaei HR, Fakhradini K, Arabzadeh SAM, Afshar AA, Iranmanesh Z, et al. (2016) Antiviral Activity of Specific siRNA against Hemagglutinin and Neuraminidase of Influenza Virus H1N1 in MDCK Cell Culture. MOJ Immunol 3(1): 00068 DOI: 10.15406/moji.2016.03.00068
H1N1 influenza virus causes respiratory syndrome on the upper respiratory system. Hemagglutinin (HA) and Neuraminidase (NA) proteins are the most important viral proteins that play a role in attachment and facilitates of releasing virus respectively. Short interfering RNA (siRNA) plays notable a roles in the RNA interference (RNAi) pathway and interferes the expression of specific genes with complementary nucleotide sequences by breaking down mRNA function and stopping transcription and translation. To explore shutting down HA and NA, siRNA were designed and genes of H1N1 are inhibited by siRNA molecules in MDCK cell culture.
The cells were infected with virus and then treated by different concentration of siRNA. The HA, NA gene expression and the viral load were investigated by Real Time PCR between 24 to 72 hours. The cells were infected with H1N1 virus at a multiplicity of infection (MOI: 3) and then treated with different concentration of siRNA (0.5–6 nmol/L) and monitored for 72 hours post transfection. Interestingly, the viral load extremely decreased at first hours, even the viral load was not detectable after 48 hours. Three nmol/L concentration of siRNA for HA was more effective. In case of siRNA for NA, 3 nmol/L concentrations shows efficacy and also cytopathic effect (CPE) was not observed in 3 nmol/L concentrations in MDCK cells. This study suggests that siRNA against NA and HA, silenced viral genes in H1N1 influenza virus infected cells. Results showed that siRNA is an efficient tool for silencing influenza virus infection. The advantage of siRNA is the specific inhibition of mRNA which can be used for inhibition of other flu viral genes and/or other viruses.
Keywords: influenza virus, h1n1, sirna, transfection, real time pcr, mutation
Influenza virus A (H1N1) is a negative single– stranded RNA virus. The RNA genome has eight segments with 13.5 kilo base pairs (kbp). Influenza virus is classified in the Orthomyxoviridae family.1,2 This virus can replicate only in living cells by binding and entering the cell. By assembly, new copies of viral proteins and RNA can make new viral particles, and exit the host cell. Viral genomes encode several proteins of which hemagglutinin (HA) and neuraminidase (NA) are the main proteins.3–4 Many animal species can be infected by Influenza virus and Birds are thought to be the main reservoirs. This virus may be transferred by aerosol droplets to human, and it attaches to host cells by binding to its receptor sialic acid in the upper respiratory system. After one or two days of incubation, common flu clinical symptoms such as fever, headache, and fatigue arise in infected patients.5 Due to its high mutation rate, a particular vaccine usually cannot offer cross protection against different serotypes.
The World Health Organization (WHO) predicts and proposes the strains of the virus which are most likely to be circulating the next year. Also the cost–effective vaccination has been widely evaluated for different groups especially in children and the elderly.6 HA and NA proteins have important roles in viral pathogenesis. SiRNA is performing as a component of RNAi pathway, which is one of the most important regulation's pathways for gene expression in animal cells.7–9 One of the antiviral defenses in vertebrates which include variety of mechanisms to inhibit virus replication is RNA silencing pathways. Gene silencing, induced by RNAi, is mediated by microRNAs (mirRNA) and small interfering RNA (siRNA) forming Watson–Crick base pairing to a target mRNA, subsequently leading to its sequence–specific cleavage.10,11
The term RNAi was first produced in the worm Caenorhabditiselegans to explain that dsRNA complementary to a particular gene was more effective at silencing the corresponding gene expression than either strand individually.12 Subsequent studies showed that silencing related to the processing of small 21–23 nucleotide–duplex short–interfering RNAs (siRNAs) and dsRNA–specific endoribonuclease III, known as Dicer, which are responsible for cleavage of dsRNA triggering the silencing machinery that produces siRNA fragments (19–23 bp duplexes). Furthermore, siRNA binds to the RNA–induced silencing complex (RISC) that leads siRNA to the mRNA.13 Then, the siRNA is unwound in an ATP–dependent manner, and the RISC complex becomes activated. Subsequently, the target mRNA is cleaved about 12 nucleotides from the 30–terminus of the siRNA strand. Small interference RNAs (siRNAs), 21–23 nucleotide long double–stranded RNA molecules could specifically cleave target's mRNAs.14
The siRNA cleavage action is not affected by the secondary structure of mRNA, and the efficiency of siRNA is determined by the siRNA specific properties.15 This feature is in contrast with the antisense, where silencing is dependent on mRNA properties such as target site accessibility, which is determined by local mRNA confirmation.16 The siRNA sequence should be 21 not long, and the sequence should preferably be 50 to 100 not downstream of the start codon, and the selected sequence site should not be in untranslated regions.17 Furthermore, the blast search of a respective organism recommended ensuring the specificity of selected siRNA sequences on the cleavage of the target sequence.18 In this study, we have designed two pair siRNA, against two important gene from influenza type A and serotype H1N1. This virus is important agent in endemic or pandemic conditions in worldwide, aim of this study shadowing two important gene from influenza H1N1 (HA, NA) and inhibition replication of the virus in cell culture.
Cell culture
The effect of siRNA on the replication of influenza virus H1N1 was measured in cell culture. The Madin–Darby canine kidney (MDCK) cells were grown in the minimum essential medium supplemented with 5% calf serum, 100 units/ml of penicillin and 100 μg/ml of streptomycin. The cells were cultured in the same type of medium without antibiotics and incubated in a 5% CO2 at 37°C for transfection. The transfection’s efficiency to determine the optimal siRNA concentration was done. Confluent 3×250 ml (75 cm2) flasks were trypsinized and cell suspensions were diluted 1:10, and then 400 μl of aliquots were transferred into each well. Before siRNA transfection, the MDCK cells were trypsinized with 5 ml of Trypsin (0.25% in media). After adding antibiotics, the cells were centrifuged at 2000 × g for 5 minutes.
The cell pellet was resuspended in 16 ml of antibiotic free medium. Cell suspension 500 μl were transferred into dishes. Before viral infection, 2 ml of medium were poured to the dishes. Cell proliferation assays were carried out in 96 well plates and MDCK cells cultured in 250 ml (75 cm2) cell culture flasks and trypsinized with 5 ml of Trypsin (0.25% in media). Cell suspension was diluted 1:66 and then 100 μl was added to each well.
H1N1 virus sample
H1N1 virus was provided by sampling via throat swap from a woman with flu signs, and viral RNA genome was extracted to confirm viral identity. The monolayer MDCK cells were placed at 370C and 5% CO2 and after appearing CPE, it was set in –700C. The infected medium was delivered to Iran national influenza center, and the identity of (H1N1) was proved. After virus selection, virus purification was done and purified virus was taken for determination of primary titer of stock virus and primary titer of virus.
Determination of virus titer
Using titration method, the highest dilution of virus (TCID50) that causes 50% of cells to show the cytopathic effect (CPE) was determined and the activity of the cells was measured with the MTT method. Statistical analyses were performed with SAS8.0 system. Also by using genome extraction kit (Bioneer, Korea) virus RNA was extracted and by Real–Time PCR (Qiagen, Germany) a virus titer of 8×106 copies/ml was determined.
Design and labeling of siRNA
For siRNA designing against HA, NA genes of influenza A virus (H1N1), gene sequences of HA,NA of the influenza virus were obtained from the NCBI web site and saved with FASTA format and by using EMBL–EBI web tools, different sequences of H1, N1 were compared. Several homologue target sites for siRNA were identified for HA, NA influenza virus. Highly effective siRNA sequences with suitable target sites selected and siRNA sense and antisense were designed by the DNA technology web tools. The siRNA was purchased from VBC Biotech (VBC–Biotech Service GmbH) and labeled by Cy5 (fluorescent dye) for being detectable after one hour of the transfection (Table 1).
|
Name |
Sequence |
|
HA -Sense |
AUGUGUAUAGCAGAACUACUA |
|
*HA-Antisense |
GUAGUUCUGCUAUACACAUUU |
|
HA-Target |
TAGTAGTTCTGCTATACACATTT |
|
NA -Sense |
UAUCUUUUGGUUUGGAUUCAU |
|
NA-Antisense |
GAAUCCAAACCAAAAGAUAAU |
|
NA-Target |
ATGAATCCAAACCAAAAGATAAT |
|
**N.C-Sense |
UUCUCCGAACGUGUCACGUdTdT |
|
N.C-Antisense |
ACGUGACACGUUCGGAGAAdTdT |
Table 1 siRNA sequence for gene silencing of the HA, NA
* labeled with Cy5 fluorescent dye **NC is negative control
siRNA transfection
PepMute™ siRNA Transfection Reagent (Signagene Laboratories) was used as novel tool for silencing gene with more than 95% efficiency. For optimal siRNA–mediated silencing, 0.5–50 nmol/L siRNA were examined and the final siRNA concentration was set more than 30 nmol/L for transfection. After preparation of monolayer MDCK cells, supernatant of medium removed 30 min prior to transfection and fresh medium was added to plates with fresh serum and antibiotic. Before infecting cells, the media removed and the cells washed two times with PBS buffer (pH=7.4) and then one milliliter of the medium ,100 µl transfection buffer, 3.6 µl PepMute™ reagent and 0.5–50 nmol/L siRNA (final concentration) were poured. To discover the maximum gene silencing, dilute siRNA and PepMute™ reagent with PepMute™ Transfection Buffer (1x) were tested.
Re–constituting siRNA stock solution should be at 10 μM, so 0.5 or 5.0 μl siRNA stock solution added per well to make final 5.0 and 50 nmol/L siRNA respectively and mixed 3.6 μl PepMute™ reagent by pipetting up and down. Incubation was done for transfection and then cells were infected with influenza virus H1N1 (MOI=3). For evaluation of silencing, transfected MDCK cells were inoculated with influenza virus H1N1 (MOI=3) and after 48 hours remove supernatant media and wash two times with PBS (pH=7.4) and the above steps are repeated. Gene silencing is usually measured 24–72 hours post transfection (Figure 1).
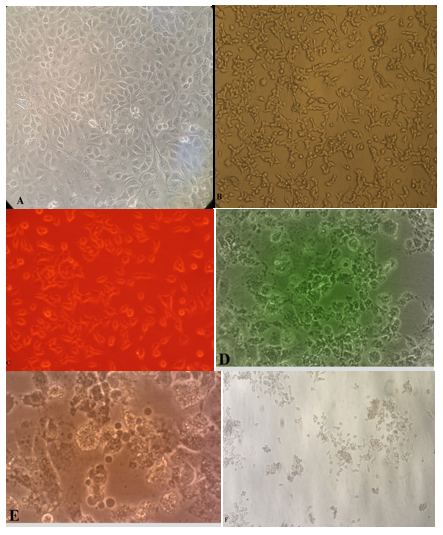
Figure 1 Effect of siRNA HA, NA transfection in MDCK cell culture. A: Normal MDCK cell line (uninfected or transected with siN.C), B: Treated MDCK cell line with siRNA HA, NA and no effect was seen after 72 hour, C: Fluorescent microscopic illustration for siRNA HA transfected MDCK cell line, D: Infected MDCK cell line with influenza H1N1 after 24 hours, E: infected MDCK cell line with influenza H1N1 after 48 hours, F: infected MDCK cell line with HSV–1 after 72 hours.
mRNA and RNA virus extraction
To evaluate silencing performance by our siRNA and replication of H1N1 after silencing of HA, NA genes, mRNA and RNA virus were extracted from cells with or without infection by RNA easy mini kit (Qiagen, Germany) and used to study HA, NA gene expression in different siRNA concentrations for evaluation of the gene silencing effects and replication of virus.
r–Real Time PCR
After extraction of mRNA and RNA virus, rRT–PCR was carried out by using cDNA synthesis kit by Revert Aid cDNA synthesis kit (Thermo scientific, USA). RNA sample was heated to 650C for 10 minutes and then transferred on ice. Based on protocol, primers and probe of kit were added. Quantities Probe PCR kit was used for Real– Time PCR (Qiagen, Germany). The eukaryotic 18s rRNA was used as internal control that was purchased from Metabion Company (Metabion international company) (Table 2).
|
Name |
Sequence |
|
HA -Forward |
AAGGGAAAGAAGTTCTCG |
|
HA-Reverse |
CTTGCTGTATCTTGATGTC |
|
HA-Probe |
FAM- CCATCCATCTACTACTGCTGACCAA-BHQ1 |
|
NA -Forward |
GGACAGACAATAACTTCTC |
|
NA-Reverse |
CCTCTGATTAGTTCAACC |
|
NA-Probe |
JOE- AGCAAGGTCTTATACAATCCAGCC-BHQ1 |
|
*18sRNA -Forward |
GGGAGGTAGTGACGAAAAATAA |
|
18sRNA-Reverse |
TTGCCCTCCAATGGATCCT |
|
18sRNA-Probe |
Texas Red- CGAGGCCCTGTAATTGGAATGAGTCCACT- BHQ1 |
Table 2 Primer and Probe sequence for determination HA and NA gene expression.
*18sRNA was used for internal control
Effect of siRNAs on viability of MDCK cell line
The activity, morphology and viability of the cells were evaluated in different concentration of siRNAs and different times after siRNA transfection by MTT method which is shown in (Figure 2 & 3). In Figure 3 the transfection of siRNA–HA, siRNA NA and siRNA–HA+NA were evaluated and the growth of MDCK cells in each group evaluated. Also 3nmol/L concentration of siRNA in period of 72 hours was studied. No harmful effect was detected and in cell+virus control.
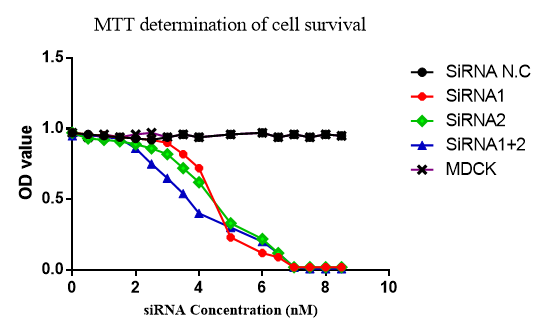
Figure 2 Effect of the different concentration of siRNA on viability of the MDCK cell line (siRNA1=HA, SiRNA2=NA, siRNA1+2= HA+NA, siRNA N.C=Negative control).
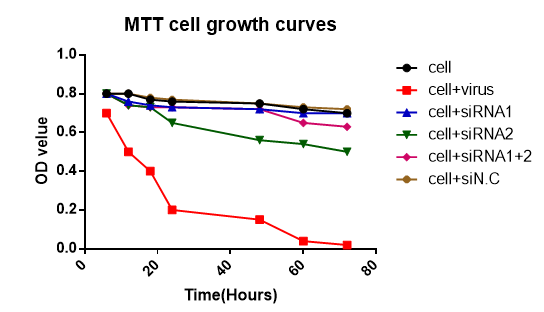
Figure 3 Effect of siRNAs in different times on viability of the MDCK cell line (siRNA1=HA, siRNA2=NA, siRNA1+2= HA+NA, siRNA N.C= Negative control).
Effect of siRNA on HA, NA gene expression
The Real–Time PCR was used to explore the effect of siRNA on mRNA translation for HA, NA proteins. For first step, different concentration of siRNAs was used to detect the optimized concentration of the siRNA in each group. The results showed that by using 3nmol/L, the HA and NA production was inhibited completely (Figure 4). In Figure 5, the decrease in the rate of HA expression was shown when the siRNA–HA was used comparing to normal cells or infected cell plus not specific siRNA (siRNA N.C) cases. In Figure 6 the rate of NA expression was decreased when the siRNA–NA used comparing to normal cells or infected cell+ not specific siRNA cases. This experiment was repeated six times and all of results indicated inhibition of HA, NA genes. At this concentration, no viral CPE was detectable and cells grow well. Compared with positive control of the virus and cell, specific siRNA–HA, siRNA–NA show inhibition in HA, NA gene expression and finally the level of viral load decreased.
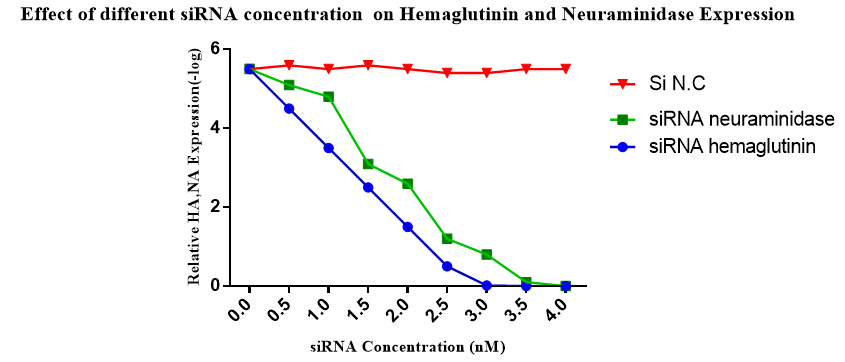
Figure 4 Effect of different siRNA concentration on gene expression of HA, NA after 72 hours post transfection.
Effect of siRNA on H1N1 virus replication
In this study, the effect of silencing on HA, NA gene expression was analyzed by siRNAs by rReal–Time PCR. Decrease in virus entrance and exit was shown in Figure 7 and finally decreases in virus efficient replication was reported. The reduction of viral titers detected in siRNA treatment and when the combination of siRNA–HA, NA used the viral load dropped faster. Comparing with positive control (cell+virus) treated with siRNA–HA, siRNA–NA indicates the reduction of viral load.
In spite of the effect of RNAi in probing gene function and in the development of novel therapeutics and antivirals, there have been only a few studies exploring the potential for RNAi against HA, NA influenza virus. These reports have focused mainly on the NP (Nucleoprotein) and M (Matrix) genes of influenza virus. In our study, we showed that influenza can be stop by the RNAi pathway.19
Both of the siRNAs showed in HA, NA gene silencing and a reduction in viral load and CPE formation. Since assays with infectious viruses are time–consuming and labor–intensive and have to be executed under elevated safety conditions, a simple method for the pre selection of active siRNA species is desirable.20 In this study, two siRNA were designed for HA and NA of influenza virus A (H1N1). The siRNAs were used and designed to especially HA, NA mRNA. To evaluate transfection performance, siRNA–NA was labeled by Cy5 and the siRNA transfection was explored by florescent microscope. siRNA has been used for several studies and have been shown that it has the potential to inhibit several genes such as viral genes. In a research which was done by Mollaie et al..12 in Iran, UL42 gene of herpes simplex virus type 1 was inhibited and virus replication decreased.21 In other research, other influenza genes like NP were inhibited by siRNA. Barik S.8 reported that clinical applications of siRNA against respiratory viruses including influenza virus, showed significant success and optimism, which was reviewed by them.9
In a study it was shown that 21 not duplexes of siRNA of the influenza virus M gene can cause specific inhibition of influenza virus matrix (M1) protein expression in transfected 293T cells. Furthermore, it was shown that a lentivirus vector can be used to effectively deliver M gene siRNAs into MDCK cells and can cause specific inhibition of M1 protein expression and influenza virus replication.20,22 In another study, three plasmid constructs expressing small interfering RNAs (siRNAs) targeted to sequences encoding the ribonucleoprotein member, nucleoprotein (NP) and/or PA, of influenza virus genome.
The antiviral properties of siRNAs against the H5N1 strain of influenza virus were studied by evaluating their capacity to silence expression of target genes as well as their effect on influenza virus–induced apoptosis in MDCK cells, chicken embryo fibroblast cells, and embryonated chicken eggs in a transient replication model. The results demonstrated that RNA interference (RNAi) can be used to inhibit protein expression and replication of influenza virus and that RNAi treatment holds potential as a new approach to prevent avian influenza.23 Small interfering RNAs (siRNAs) specific for conserved regions of the viral genome can potently inhibit influenza virus production in cell lines, and BALB/c mice. siRNA expression plasmid pBabe–Super was chosen in this study, which directed the synthesis of small interfering RNAs in cells. The findings reveal that newly synthesized nucleocapsid, polymerase A (PA) and polymerase B1 (PB1) proteins are required for avian influenza virus transcription and replication and provide a basis for the development of small interfering RNAs as prophylaxis and therapy for avian influenza infection in birds and humans.24
Authors suggested that pulmonary delivery of siRNA has the considerable therapeutic potential for treating viral respiratory infectious diseases, including influenza. By introducing siRNA that targets the critical region of viral genes, viral mRNAs can be degraded and viral replication can be inhibited in mammalian cells. To enable siRNA to be used as an antiviral agent, the nucleic acid delivery barrier must be overcome. Effective local delivery of siRNA to lung tissues is required to reduce the therapeutic dose and minimize systemic adverse effects. To develop a formulation suited for clinical application, complexes of pH–responsive peptides, containing either histidine or 2,3–diaminopropionic acid (Dap), and siRNA was prepared into dry powders by spray drying with mannitol, which was used as a bulking agent.
The spray–dried (SD) powders were characterized and found to be suitable for inhalation with good stability, preserving the integrity of the siRNA as well as the biological and antiviral activities. The formulations mediated highly effective in vitro delivery of antiviral siRNA into mammalian lung epithelial cells, leading to significant inhibition of viral replication when the transected cells were subsequently challenged with the H1N1 influenza virus. SD siRNA powders containing pH–responsive peptides are a promising inhalable formulation to deliver antiviral siRNA against influenza and are readily adapted for the treatment of other respiratory diseases.25,26 Our crucial result is that the siRNA designed here is specified for the target region (HA, NA of Influenza A) with high efficiency. The desired characteristics of these molecules are their special performance and non–harmful effects. Our siRNA is an important tool which is able to act as the non–viral agent with no complication for cells and act specially and also have acted well in vitro and showed considerable results.
The evolution of siRNA–mediated antiviral therapy has the greatest benefit among populations infected with virus strains resistant to routine antivirals and in cases of severe or recrudescent disease. The in vitro system depicted will allow us to more easily follow up further studies of siRNA transfection and provides a foundation for the development of in vivo applications of siRNA. In vivo activity of RNAi has been demonstrated for both viral and host genes.
These data suggest that siRNA can be developed as a drug. In this survey, specific siRNA against HA, NA proteins of influenza A virus (H1N1) were used and they induce a sharp decrease in viral load in some concentrations. Interestingly, the level of CPE virus was not observable and cells had complete normal life. It is suggested a continued research on use of siRNA and nanoparticles for delivery siRNA into the cells against respiratory viruses, because these viruses cause worldwide mobility and mortality. At present, studies are underway on proteomics and genomics effects of the siRNA transfection in other respiratory infections.
The authors of this project are grateful to Kerman Virology Laboratory in Faculty of medicine staff and their cooperation in cell culture, National influenza research center in Tehran for prepared type of virus, and so thanks for physiology research center ethics committee of the Kerman University of Medical Sciences in approve this project.

©2016 Mollaei, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.