MOJ
eISSN: 2373-4442


Research Article Volume 1 Issue 3
1Department of Pathology, College of Medicine, University of Arkansas for Medical Sciences, USA
2Department of Microbiology and Parasitology, China Medical University, China
3Department of Obstetrics & Gynecology, College of Medicine, University of Arkansas for Medical Sciences, USA
Correspondence: Mayumi Nakagawa, Department of Pathology, College of Medicine, University of Arkansas for Medical Sciences, 4301 West Markham Street, Little Rock, AR 72205, USA, Tel 501-686-8635, Fax 501-526-4621
Received: August 09, 2014 | Published: August 9, 2014
Citation: Coleman HN, Wang X, Greenfield WW, Nakagawa M (2014) A Human Papillomavirus Type 16 E6 52-62 CD4 T-Cell Epitope Restricted by the HLA-DR11 Molecule Described in an Epitope Hotspot. MOJ Immunol 1(3): 00018. DOI: 10.15406/moji.2014.01.00018
Cell-mediated immune responses to the human papillomavirus type 16 (HPV 16) E6 protein have been shown to be important in viral clearance and in regression of cervical lesions. Here, detailed analyses of a novel HPV 16 E6 CD4 T-cell epitope from a subject with cervical intraepithelial neoplasia 1 are described. This subject had demonstrated HPV 16 CD4 T-cell responses to multiple regions within the E6 protein. Isolation and cloning of CD4 T-cells were performed by magnetic selection of interferon-secreting cells and limiting dilution. A single HPV 16-specific T-cell clone isolated was shown to have a specificity to HPV 16 E6 52-62 restricted by the HLA-DR11 molecule. Homologous sequences (≥70% amino acid homology) were identified for HPV types 31, 33, 45, 58, 73, but cross-recognition was demonstrated only for HPV 45. Based on work performed by our group and others, it is known that this short peptide contains multiple CD4 and CD8 T-cell HPV epitopes and would be an ideal region to incorporate into a design of vaccines and immunotherapies against HPV-associated malignancies.
Keywords:human papillomavirus, cd4; t-cells, epitope, hotspot
HPV 16, human papillomavirus type 16; HPV, human papillomavirus; IFN-γ: interferon-γ; ELISPOT, enzyme-linked immunospot Pap: papanicolaou; CIN, cervical intraepithelial neoplasia; PBS, phosphate-buffered saline; LCL, B-lymphoblastoid cell line
Cervical cancer cases are almost always associated with human papillomaviruses (HPVs). They can also cause other malignancies including anal, oropharyngeal, penile, vaginal, and vulvar cancers, and are estimated to be responsible for 5.2% of all cancer cases globally.1,2 There are approximately 120 types of HPVs described to date,3 and a few dozen are considered to be high-risk or associated with malignancy.4 Among these high-risk HPV types, human papillomavirus type 16 (HPV 16) is the most common and is associated with approximately half of cervical cancer cases worldwide.4-6
The importance of T-cell responses against HPV 16 E6 protein in clearance of HPV infection was first demonstrated by examining T-cell responses using chromium release assay and interferon-γ (IFN-γ) enzyme-linked immunospot (ELISPOT) assay in a cohort of young women being studied for natural course of HPV infection.7,8 Further studies of women with abnormal Papanicolaou (Pap) smear results also demonstrated the important of CD49 and CD810 T-cell responses to the E6 protein in regression of the cervical lesions. In this manuscript, a novel CD4 T-cell epitope, HPV 16 E6 52-62 peptide restricted by the HLA-DR11 molecule, is described through detailed analyses of CD4 T-cell responses of one subject with cervical intraepithelial neoplasia (CIN) 1.
Subject
The subject was selected among other participants of previously published study [9] for further characterization of HPV 16-specific CD4 T-cell response for having demonstrated positive CD4 T-cell responses to multiple HPV 16 E6 regions (Figure 1). The study was approved by the institutional review board, and written informed consent was obtained.

Figure 1 An ELISPOT assay for identifying regions containing potential HPV-specific CD4 T-cell epitopes. One hundred thousand CD4 T-cells stimulated in vitro with autologous dendritic cells pulsed with HPV 16 E6 and E7 proteins were tested using overlapping peptides.9 Each peptide pool contains three overlapping 15-mer peptides. “*” indicates regions with positive CD4 T-cell responses defined by having spot forming units ≥2 times those in the media only control wells. The bars represent standard error of means.
HPV-DNA testing
The Linear Array HPV Genotyping Test which detects 37 anogenital HPV types [6, 11, 16, 18, 26, 31, 33, 35, 39, 40, 42, 45, 51, 52, 53, 54, 55, 56, 58, 59, 61, 62, 64, 66, 67, 68, 69, 70, 71, 72, 73 (MM9), 81, 82 (MM4), 83 (MM7), 84 (MM8), IS39, and CP6108] was used according to manufacturer’s instructions (Roche Diagnostics, Indianapolis, IN).
Peptides
HPV 16 peptides ranging from 8 to 15 amino acids long were synthesized based on sequences from the HPV 16 German prototype.11 Homologous peptides 15 amino acids long in length from other high-risk HPV types [4] were identified and synthesized for having ≥70% amino acid homology. The purities of peptides ranged from 70 to 90%.
Magnetic selection of IFN-γ secreting antigen specific T-cells and T-cell cloning
The peptides (10 µM each) included in three HPV 16 E6 regions with the most robust CD4 T-cell responses (HPV 16 E6 16-40, 46-70, and 91-115) were used to stimulate CD4 T-cells for 3 to 4 hours, and antigens-specific IFN-γ secreting cells were selected using IFN-γ Secretion Assay Cell Enrichment and Detection Kit (Miltenyi Biotec Inc, Auburn, CA, USA). Selected cells were plated with feeder cell mixture as previously described to isolate T-cell clones.12,13 Molecular analysis to formally establish the clonality was not performed.
Screening T-cell clones
One hundred µl of culture medium containing the T-cell clones were collected, washed, and tested in an ELISPOT assay12 with the peptides covering the HPV 16 E6 16-40, 46-70, 91-115 regions (Table 1). A corresponding negative control contained media. First, 96-well plates (MultiScreen; Millipore, Bedford, MA) were coated with primary anti-IFN-γ monoclonal antibody, 1-DIK (Mabtech, Stockholm, Sweden) at a concentration of 5µg/ml overnight at 4 °C. The plates were washed with phosphate-buffered saline (PBS) and were blocked with 50µl of RPMI 1640 medium supplemented with 5% human serum for 1 hour at 37°C. One-hundred thousand autologous Epstein-Barr Virus-transformed B-lymphoblastoid cell line (LCL) cells were added to each well. Fiftyµl of media containing T-cell clone cells was plated to one well at the same position in duplicate (to test the peptide pool and media). The final concentration of each peptide was 10 µM. One phytohaemagluttinin (10µg/ml) positive control well and one no T-cell clone negative control well was set on each plate. After 20 hour incubation, wells were washed with PBS containing 0.05% Tween-20. Biotin-conjugated anti-IFN-γ monoclonal antibody (1µg/ml, Mabtech) was added, and the plates were incubated for 2 hours at 37 °C. After washing the plate with PBS containing 0.1% Tween-20, Avidin-bound biotinylated horseradish peroxidase H (Vectastain Elite Kit; Vector laboratories, Inc., Burlingame, CA), the plates were incubated for 1hour at 37 °C. Wells were washed with PBS containing 0.1% Tween-20 and were developed using stable diaminobenzene (Life Technologies, Carlsbad, CA) at room temperature. The plates were washed with deionized water and were air-dried. Spot-forming units were counted using an automated ELISPOT analyzer (AID ELISPOT Classic Reader; Autoimmun Diagnostika GmbH, Strassberg, Germany). The wells that showed spots in an ELISPOT plate with the peptide pool, but not in the other media plate, were considered to contain potential T-cell clones with the specificity of interest. These T-cell clones were further expanded.12
Confirming the specificity of T-cell clones
Screening positive T-cell clones were tested again using three separate peptide pools covering the HPV 16 E6 16-40, 46-70, 91-115 regions respectively. Then, the 15-mer peptides (10µM each) contained in the positive region were tested individually. One thousand T-cell clone cells were seeded along with 1x105 autologous LCL cells per well in duplicate and ELISPOT assay was performed as described above.
Identifying the core amino acid sequence of the novel HPV 16 E6 CD4 epitope
Serial ELISPOT assays were performed using HPV 16 E6 peptides ranging from 8 to 15 amino acids in length (Table 1) as described above. A single peptide concentration of 10µM (Figure 2) and concentrations over a 4 log range (Figure 3) were used.

Figure 2 An ELISPOT assay for defining the core sequence of the novel HPV 16 E6 epitope. The bars represent standard error of means.
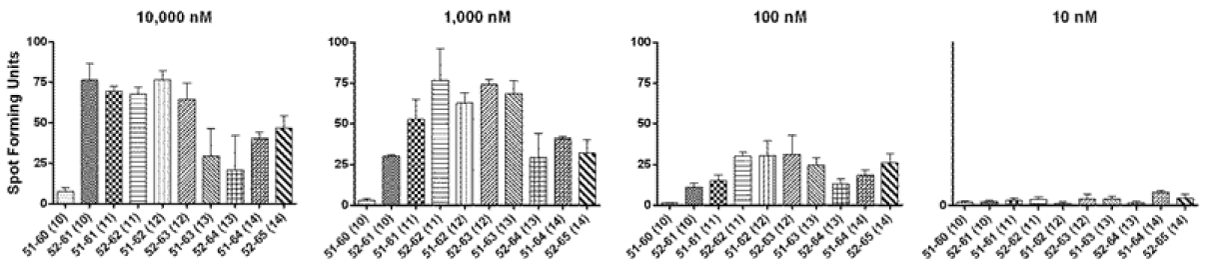
Figure 3 An ELISPOT assay for defining the core sequence of the novel HPV 16 E6 epitope using serially diluted peptides. A representative of two experiments is shown. The bars represent standard error of means.
Identifying the restriction HLA class II molecule
Seven allogeneic LCL cells sharing one, two or no class II molecule(s) with the subject being studied (HLA-DQ7, -DQ8, -DR4, - DR11) were utilized for ELISPOT assay in order for identifying the restriction molecule of the epitope being described (Figure 4) [14]. One thousand T-cell clone cells were plated along with 1 x 105 alloegeneic LCL cells per well in duplicate. The HPV 16 E6 51-65 (15-mer) peptide was added at 10µM along with 20U/ml of rhIL-2.
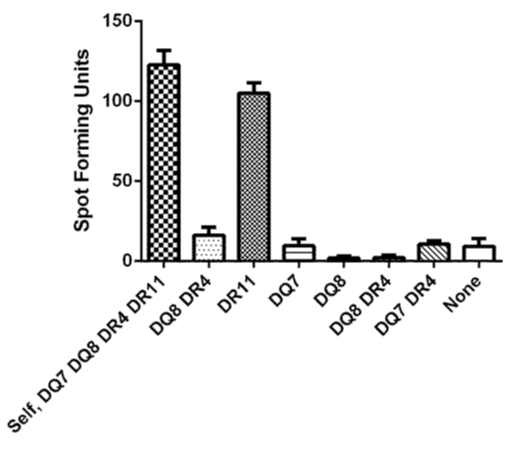
Figure 4 An ELISPOT assay defining the HLA class II restriction element for the novel HPV 16 E6 epitope. Autologous LCL and allogeneic LCLs sharing none to two class II HLA molecules were used. A representative of two experiments performed is shown. The bars represent standard error of means.
Recognition of homologous CD4 T-cell epitopes from other high-risk HPV types
Recognition of homologous epitopes from other high-risk types [HPV 31 44-58 (15-mer), HPV 33 44-58 (15-mer), HPV 45 46-60 (15-mer), HPV 58 44-58 (15-mer), and HPV 73 45-59 (15-mer), 10 µM each] were assessed using an ELISPOT assay (Figure 5).
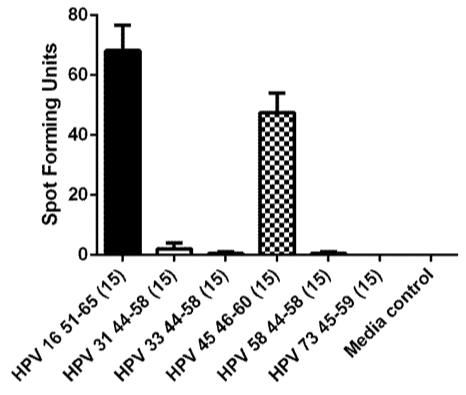
Figure 5 An ELISPOT assay assessing the cross-recognition of homologous peptides from other high-risk HPV types. Homologous peptides from HPV 31, 33, 45, 58, and 73 which had ≥70% amino acid homology with the HPV 16 51-65 sequence were synthesized and tested (Table 3). The experiment was performed twice. The bars represent standard error of means.
HLA typing
HLA typing was performed at the University of Arkansas for Medical Sciences HLA laboratory using peripheral blood lymphocytes or LCLs using PCR sequence-specific amplification method.
Fluorescence-activated cell sorter analysis
The surface phenotype of the T-cell clone was determined by staining with CD3-fluorescein isothiocyanate, CD8-phycoethythrin, CD4-PerCP-Cyanine 5.5, and CD56 (NCAM)-allophycocyanin (eBioscience, San Diego, CA). Isotype-matched antibodies were used as a negative control. Data were acquired and using FACS Fortessa and analyzed using FACS Diva (BD Biosciences, San Jose, CA) at the University of Arkansas for Medical Sciences Microbiology and Immunology Flow Cytometry Core Laboratory.
The subject was a 29 year old female enrolled in a previously published study examining HPV-specific CD4 T-cell responses in women being followed for abnormal Pap smear results [9]. Four months prior to the date of enrollment, the patient had a Pap smear which showed low grade squamous intraepithelial lesion. On the enrollment day, a blood sample was drawn for performing ELISPOT assays using CD4 T-cells examining responses to the HPV 16 E6 and E7 proteins using synthetic peptides, and positive responses were demonstrated in multiple E6 regions (Figure 1). A thin-prep sample was collected for HPV-DNA testing which was positive for HPV types 39 (high-risk), 54 (low-risk), and 82 (high-risk). Colposcopy guided biopsy performed on the day of enrollment showed CIN 1. However, the follow up Pap smear performed 6 months later was normal.
Attempts were made to isolate T-cell clones with specificities from three HPV 16 E6 regions (16-40, 46-60, and 91-115) to which the highest CD4 responses were demonstrated (Figure 1) using overlapping peptides covering these regions (Table 1). One hundred nine T-cell clones were isolated after IFN-γselection and limiting dilution. Screening ELISPOT assay using peptides shown in Table 1 identified 5 potential HPV-specific T-cell clones. These 5 T-cell clones were tested using pools of peptides covering E6 16-40, E6 46-70, or E6 91-115, and one of them demonstrated specificity to the E6 46-70 pool. The other 4 T-cell clones were false positives. Testing this T-cell clone using the three peptides in the pool individually narrowed the specificity to the E6 51-65 region.
HPV Type |
Region |
Amino Acid Residues |
Sequence |
16 |
E6 16-40 |
E6 16-30 (15-mer) |
PRKLPQLCTELQTTI |
16 |
E6 16-40 |
E6 21-35 (15-mer) |
QLCTELQTTIHDIIL |
16 |
E6 16-40 |
E6 26-40 (15-mer) |
LQTTIHDIILECVYC |
16 |
E6 46-70 |
E6 46-60 (15-mer) |
RREVYDFAFRDLCIV |
16 |
E6 46-70 |
E6 51-65 (15-mer) |
DFAFRDLCIVYRDGN |
16 |
E6 46-70 |
E6 56-70 (15-mer) |
DLCIVYRDGNPYAVC |
16 |
E6 91-115 |
E6 91-105 (15-mer) |
YGTTLEQQYNKPLCD |
16 |
E6 91-115 |
E6 96-110 (15-mer) |
EQQYNKPLCDLLIRC |
16 |
E6 91-115 |
E6 101-115 (15-mer) |
KPLCDLLIRCINCQK |
Table 1 A list of peptides used to isolate IFN-γ secreting antigen-specific T-cells
In order to define the core sequence of this HPV 16 E6 epitope (i.e. the peptide minimal in length that shows optimal response), peptides from the E6 51-65 regions that were 8, 9, 10, or 11 amino acids long in length (Table 2) were tested using an ELISPOT assay (Figure 2). Positive responses to HPV 16 E6 51-60 (10-mer), 52-61 (10-mer), 51-61 (11-mer), 52-62 (11-mer), and 51-65 (15-mer) were shown. Then, the peptides 10 to 14 amino acids long in lengths were serially diluted 10 fold and were tested at 10,000 nM, 1,000 nM, 100 nM, and 10nM (Figure 3). Although E6 52-61 (10-mer), 51-61 (10-mer), and 52-62 (11-mer) peptides had similar levels of positivity at 10,000 nM, the 52-62 (11-mer) peptide retained the highest reactivity at 1,000 nM and 100 nM. Therefore, the core sequence of this novel HPV 16 E6 epitope appears to be HPV 16 E6 52-62 (11-mer).
HPV Type |
Region |
Amino Acid Residues |
Sequence |
16 |
E6 51-65 |
E6 51-65 (15-mer) |
DFAFRDLCIVYRDGN |
16 |
E6 51-65 |
E6 51-64 (14-mer) |
DFAFRDLCIVYRDG |
16 |
E6 51-65 |
E6 52-65 (14-mer) |
FAFRDLCIVYRDGN |
16 |
E6 51-65 |
E6 51-63 (13-mer) |
DFAFRDLCIVYRD |
16 |
E6 51-65 |
E6 52-64 (13-mer) |
FAFRDLCIVYRDG |
16 |
E6 51-65 |
E6 51-62 (12-mer) |
DFAFRDLCIVYR |
16 |
E6 51-65 |
E6 52-63 (12-mer) |
FAFRDLCIVYRD |
16 |
E6 51-65 |
E6 51-61 (11-mer) |
DFAFRDLCIVY |
16 |
E6 51-65 |
E6 52-62 (11-mer) |
FAFRDLCIVYR |
16 |
E6 51-65 |
E6 53-63 (11-mer) |
AFRDLCIVYRD |
16 |
E6 51-65 |
E6 54-64 (11-mer) |
FRDLCIVYRDG |
16 |
E6 51-65 |
E6 55-65 (11-mer) |
RDLCIVYRDGN |
16 |
E6 51-65 |
E6 51-60 (10-mer) |
DFAFRDLCIV |
16 |
E6 51-65 |
E6 52-61 (10-mer) |
FAFRDLCIVY |
16 |
E6 51-65 |
E6 53-62 (10-mer) |
AFRDLCIVYR |
16 |
E6 51-65 |
E6 54-63 (10-mer) |
FRDLCIVYRD |
16 |
E6 51-65 |
E6 55-64 (10-mer) |
RDLCIVYRDG |
16 |
E6 51-65 |
E6 56-65 (10-mer) |
DLCIVYRDGN |
16 |
E6 51-65 |
E6 51-59 (9-mer) |
DFAFRDLCI |
16 |
E6 51-65 |
E6 52-60 (9-mer) |
FAFRDLCIV |
16 |
E6 51-65 |
E6 53-61 (9-mer) |
AFRDLCIVY |
16 |
E6 51-65 |
E6 54-62 (9-mer) |
FRDLCIVYR |
16 |
E6 51-65 |
E6 55-63 (9-mer) |
RDLCIVYRD |
16 |
E6 51-65 |
E6 56-64 (9-mer) |
DLCIVYRDG |
16 |
E6 51-65 |
E6 57-65 (9-mer) |
LCIVYRDGN |
16 |
E6 51-65 |
E6 53-60 (8-mer) |
AFRDLCIV |
16 |
E6 51-65 |
E6 54-61 (8-mer) |
FRDLCIVY |
Table 2 A list peptides used to describe the core sequence of the novel HPV 16 E6 epitope
The surface phenotype of this clone was shown to be CD3+CD4+CD8-CD56- by flow cytometry confirming it is a CD4 T-cell clone (data not shown). An ELISPOT assay using allogeneic LCLs sharing one, two, or no HLA class I type(s) with the subject showed that the restriction element for this CD4 T-cell epitope is HLA class II DR11 (Figure 3).
Of 15 high-risk HPV types described by Munos et al. [4], 5 of them had homologous E6 peptide with ≥70% amino acid homology to the HPV 16 E6 51-65 (15-mer) peptide (Table 3). The CD4 T-cell clone was able to cross-recognize the homologous peptide from HPV 45 (Figure 4).
HPV Type |
Amino Acid Residues |
Sequence |
16 |
E6 51-65 (15-mer) |
DFAFRDLCIVYRDGN |
31 |
E6 44-58 (15-mer) |
DFAFTDLTIVYRDDT |
33 |
E6 44-58 (15-mer) |
DFAFADLTVVYREGN |
45 |
E6 46-60 (15-mer) |
QFAFKDLCIVYRDCI |
58 |
E6 44-58 (15-mer) |
DFVFADLRIVYRDGN |
73 |
E6 45-59 (15-mer) |
DFAFSDLCIVYRDKP |
38 |
E6 46-60 (15-mer) |
EFAFSDLYVVYRDGE |
54 |
E6 40-54 (15-mer) |
PRTLADLCKVCNIPM |
82 |
E6 44-58 (15-mer) |
NVAFTELRIVYRDNT |
Table 3 A list of peptides from high-risk HPV types with ≥70% homology with HPV 16 E6 51-65 sequences, which were used to assess cross-recognition. The sequences with <70% homology from HPV types 38, 54, and 82, which were detected in the subject, are also shown.
Amino acid residues different from the HPV 16 are shown in bold
T-cell responses to the HPV 16 E6 protein have been shown to be important in viral clearance7,8 and regression of cervical lesions.9,10 This manuscript describes detailed analyses of CD4 T-cell responses to HPV 16 E6 protein of a 29 year old woman with LSIL who had participated in our clinical study.9 An ELISPOT using CD4 T-cells demonstrated multiple positive regions (Figure 1). After attempting to isolate T-cell clones from the three most immunogenic regions (E6 16-40, 46-70, and 91-115), a single T-cell clone recognizing an epitope in the E6 46-70 region was isolated and characterized. Surface phenotyping and fine epitope mapping revealed a novel CD4 epitope with the core sequence of HPV 16 E6 52-62 (11 amino acid long). On the day the blood sample was drawn from which this T-cell clone was isolated, the subject had persistent CIN 1 diagnosed with a biopsy, and was positive for HPV types 39, 54, and 82. Therefore, the detected HPV 16-specific CD4 T-cell responses were likely directed to past HPV 16 infection as the presence of circulating HPV 16-specific T-cells years after the HPV 16 infections had become undetectable has been clearly demonstrated.15,16 Furthermore, HPV 16 is the most common HPV type detected.4-6,17,18 The cross-recognition of HPV 39, 54, or 82-specific T-cells against the HPV 16 E6 52-65 sequence seems unlikely as peptide sequences from HPV 39, 54, and 82 in this E6 region have low homology (Table 3).
HPV 16 E6 52-62 CD4 T-cell epitopes restricted by HLA-DR11 has not been described previously to our knowledge. Interestingly, another epitope E6 52-61, within in the new epitope, restricted by DP0201 has been described. It is not known whether the E6 52-61 (10-mer) is the optimal sequence for this epitope since peptides of various lengths were not tested; however, this epitope was shown to be endogenously processed and presented since the T-cells recognized antigen presenting cells pulsed with the whole HPV 16 E6 protein.19 The HPV 16 E6 52-62-specific T-cell clone we describe here can also recognize the E6 52-61 peptide (Figure 2 and 3); therefore, the E6 52-62 epitope is likely to be endogenously processed and presented through the same MHC class II pathway.
The HPV 16 E6 52-61epitope is also a CD8 T-cell epitope restricted by B35,12 B57,20 and B58.13 Therefore, this region contains multiple CD4 and CD8 epitopes, and would be ideal for vaccine development as well as immunotherapy since the efficacy of treating HPV-associated tumor using a peptide containing CD4 and CD8 T-cells epitopes have been demonstrated.21 Furthermore, this region of the E6 protein is well conserved among high-risk HPV types, and homologous peptides from HPV 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, and 73 have been tested for cross-reactivity.13,22 The CD8 T-cell clone restricted by B57 had significant cross-reactivity (≥50% of spot-forming units compared to HPV 16 peptide) with homologous peptides from HPV 35, 39, 45, 51, and 7322 while the CD8 T-cell clone restricted by B58 cross-recognized homologous peptides from HPV 31, 33, 35, 39, 45, 51, 58, and 73.13 In the current work, a homologous peptide from HPV 45 (Table 3) was also shown to be recognized by the HPV 16 E6 52-62-specific CD4 T-cell clone. However, whether or not such cross-recognition is present in natural infection is not known as whether the homologous epitopes are generated from the native protein by natural antigen processing has not been investigated. Nevertheless, HPV 16 E6 52-62 region contains multiple CD4 and CD8 T-cell epitopes. Such an immunologically active region should be incorporated into therapeutic strategies to prevent and/treat cancer cases caused by high-risk HPV.
A novel HPV 16 E6 52-62 CD4 T-cell epitope restricted by the HLA-DR11 molecule is described. This small region contains multiple CD4 and CD8 T-cell epitopes making it ideal for inclusion in vaccines and immunotherapy’s against HPV-associated malignancies.
This study was supported by grants from the National Institutes of Health (R01CA143130, UL1TR000039, and P20GM103625).
Although not directly related to the work described in this manuscript, Mayumi Nakagawa is an inventor named in the patent (#8,652,482) and patent applications (US Serial #s 13/136,557, 61/887,425, 61/890,130, and 61/890,306) describing a peptide-based HPV therapeutic vaccine. Other authors do not have any potential conflict of interests to declare.

©2014 Coleman, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.