MOJ
eISSN: 2373-4442


Research Article Volume 5 Issue 5
1College of Chemistry, Chemical Engineering and Biotechnology, Donghua University, Shanghai, 00, China.
2Department of Biological Sciences, Faculty of Science, King Abdul Aziz University, Saudi Arabia
Correspondence: Fariha Kanwal, College of Chemistry, Chemical Engineering and Biotechnology, Donghua University, Shanghai, 201620, China, Tel 862-167-792-740
Received: January 01, 1971 | Published: June 14, 2017
Citation: Kanwal F, Simair AA, Chen T, Zhang YL, Qadri I, et al. (2017) Molecular Cloning and In Silico Analysis of hIL-6 Gene from Pakistani Dengue Hemorrhagic Fever Patients. MOJ Immunol 5(5): 00170. DOI: 10.15406/moji.2017.05.00170
Cytokines are the vital participating agents to regulate the immune system. They conclude both the categories and degree of an immune response generated after any infection or vaccination. Dengue virus (DV) has been endemic for many years in Pakistan with co-circulating cross-reactive serotypes. It shows up-regulation of hIL-6 (human Interleukin-6) level rendering this cytokine as an essential element to discover DV pathogenesis and therapeutic potential for severe DV infection. In this study, the hIL-6 gene was amplified and cloned into pCR®2.1 vector from Pakistani normal healthy individuals, DF (Dengue fever) and DHF (Dengue hemorrhagic fever) patient’s serum, following an in silico sequence analysis of transformed gene. The gene in recombinant pCR®2.1/hIL-6 was sequenced and submitted to GenBank (Accession No. JQ250825.1). In silico analysis of the DHF serum driven hIL-6 gene revealed a non-significant deletion of Glycine at position 17 in a protein sequence. Further sequence analysis of up-regulated hIL-6 gene through predicted secondary and 3-D model indicates no functional oddity with 99% identity to its structural homologs. Additional structural regulations and expression level of hIL-6 studies will aid to understand its role in DV pathogenesis and therapeutic approaches towards severe dengue infection.
Keywords: human interleukin-6, dengue, DHF, molecular cloning, cytokine,In silico analysis
DV, dengue virus; DHF, dengue hemorrhagic fever; IL-6, interleukin-6; Hil-6, human interleukin-6
Interleukin 6 (IL-6) is an important player against infections and tissue damage in mammalian host defense. Its biological functions are evidently spacious including hematopoiesis, modulation of immune response, regulation of endocrine and nervous systems, protective plasma proteins, acute-phase proteins by the liver and control of body temperature.1–3 IL-6 works in an expansive manner on cells of the immune and non-immune system and most often behave like a hormone affecting homeostatic processes. Its prompt induction starts with the onset of any tissue damage or inflammation, activating the host defense and terminates upon tissue homeostasis restoration.4,5 However, the deregulated persistent synthesis of IL-6 has been seen in the development of various autoimmune, chronic inflammatory diseases and cancers.6 Clinical trials using the humanized anti-IL-6 receptor monoclonal antibody have verified the efficacy of IL-6 blockade for the treatment of unmanageable inflammatory diseases, such as systemic juvenile idiopathic arthritis, rheumatoid arthritis, and Castleman’s disease. Its situation-dependent pro and anti-inflammatory nature makes cytokine as a top intention for clinical intervention.7–10 However, IL-6 have several alternative molecular forms with multi-pattern post-translational modifications but its diverse cellular response is unclear and the recombinant IL-6 from bacteria without modifications reproducing all known functions left these post-translational modifications as non-causative for diverse actions of IL-6.11,12
Pleiotropic nature of this cytokine is clear as it has been studied under different names (hybridoma/plasmacytoma growth factor, B-cell stimulatory factor-2, IFN-b2, 26-kDa protein and HSF) at different times.3,13,14 DV infection is an arboviral disease in humans caused by a Flaviviridae family virus (DV) and this infection has an elevated worldwide impact of around 3.9 billion in both tropical and sub-tropical areas. The incidence of dengue infection got a steady rise of 30-fold during the past 50 years.15,16 Cytokine IL-6 is one of the top candidates as a prognostic marker for severe DV infection because its upregulation has been reported through the course of severe infection.17,18 Here we have attempted to determine any structural change in hIL-6 gene overproduced during DHF based on molecular cloning and sequence comparison. This work shows that the hIL-6 gene sequence clones from DHF patient’s serum are 99% identical to its structural homologs. Further structural studies of hIL-6 may help to block its continual synthesis during DHF ultimately leading towards the development of more specific preventive or therapeutic approaches.
RNA extraction, cDNA synthesis and PCR amplification
RNA was extracted from 5 normal healthy controls, 5 DF, and 11 DHF patient's blood in a 5 mL K3 Ethylene Diamine Tetra Acetate (EDTA) tubes (BD Vacutainer TM, USA) after the informed written consent from patient/guardian. The blood was immediately processed for RNA extraction using TRIZOL reagent. RNA yield was determined through Nanodrop (Eppendorf Biophotometer Plus) at 260nm. Purity was determined by calculating the ratio of absorbance at 260 nm to 280 nm. Samples having a 260/280 ratio of 1.7-2.0, and concentration of ≥100 ng/μL were subjected to cDNA synthesis. cDNA was synthesized through Revert Aid first strand cDNA synthesis kit (Fermentas). Sense and anti-sense primers for the hIL-6 gene were designed and the gene was amplified by Polymerase Chain Reaction (PCR) using the following primers resulting in the product of ~640 bp.
Sense primer: 5’AAGTCGACATGAACTCCTTCTCCACAAGCG3’Antisense primer: 5’GGAAGCTTCATTTGCCGAAGAGCCCTCA3’PCR was performed on ESCO Thermal Cycler system (Swift MaxPro, Singapore) by using Taq DNA polymerase (Thermo Scientific, USA). cDNA was denatured for 5 minutes at 95˚C followed by 35 cycles of further denaturation at 94˚C for 45 seconds, annealing at 67˚C for 60 seconds and extension at 72 ˚C for 2 minutes. A final extension of 72˚C was also carried out for 10 minutes after 35 cycles. Amplified DNA product was analyzed through 1% agarose gel in TAE buffer. DNA bands were visualized and an image was saved at gel documentation system (Wealtec, Dolphin-Doc, USA).
TA cloning and confirmation
The gel purified hIL-6 PCR product was cloned into pCR®2.1 vector (Invitrogen) provided with TA Cloning Kit and transformed into E. coli Top10 competent cells. Recombinant clones were selected based on blue/white screening. White colonies were tested for tentative positive hIL-6 clones and were confirmed by restriction digestion analysis and sequencing for~640bp size. The recombinant plasmid was isolated from suspected positive white colonies by alkaline lysis method and double digested using restriction enzymes SalI and HindIII. Plasmids isolated by alkali denaturation method were sent for sequencing. Sequencing of cloned hIL-6 gene was performed through CEQ8000 Genetic Analysis System (Beckman Coulter, USA) by using universal M13 reverse and forward primers.
In silico analysis of hIL-6 gene
The hIL-6 gene sequence was analyzed through different bioinformatics tools to evaluate its structure. The nucleotide sequence of hIL-6 gene was aligned with available reported sequences using CLC workbench. Based on the nucleotide sequence the hIL-6 protein sequence was predicted using ExPASy Translate tool. The secondary and 3-D structures were predicted to figure out any significant change in the protein sequence. Secondary structure was predicted using bioinformatics tool PSIPRED (Protein secondary structure prediction server) and 3-D model was attained through protein thread modeling approach by employing the web-based tool i-TASSER, which uses three consecutive steps of threading, fragment assembly and iteration. The accuracy of the model was evaluated by checking stereo chemical properties through PROCHECK. Similarity assessment of the predicted protein structure was checked through PROSESS, a web-based interface. ERRAT program from NIH server was used to analyze the statistics of non-bonded interactions between different atom types. It is a protein structure verification algorithm that is well suited to evaluate the progress of crystallographic model building and refinement. ERRAT method is sensitive to smaller errors than 3-D Profile analysis19 but is more lenient than PROCHECK.20 The models were visualized using SPDBV and POLYVIEW 3-D.
Cloning
The hIL-6 gene from a healthy control, DF and DHF patients' blood serum was amplified through PCR, following RNA extraction and cDNA synthesis as described in experimental section. DNA product of ~640bp was amplified by using given primers and the resultant product was analyzed through 1% agarose gel as shown in Figure 1. Lane M was representative of 1000-bp DNA Marker and Lane 1 was showing the PCR amplified hIL-6 gene (~640bp) from normal healthy control, Lane 2-3 from DF and Lane 4-5 from DHF patient serum sample. The gel purified hIL-6 PCR products were cloned into pCR®2.1 vector (TA) and transformed into E. coli Top10 competent cells. Recombinant clones were selected based on blue/white screening. White colonies were tentative positive clones for the hIL-6 fragments and were replica plated and incubated overnight at 37 °C. Confirmation was done through restriction digestion analysis and sequencing for ~640bp size. Recombinant plasmids were isolated from suspected positive white colonies by alkaline lysis method. The plasmids were double digested using restriction enzymes SalI and HindIII, releasing the fragment from the plasmid in positive clones. Two bands ~3.9 kb corresponding to TA cloning vector and ~640 bp corresponding to hIL-6 gene were observed through agarose gel as shown in Figure 2. Lane M was representative of 1000-bp DNA Marker and Lane 1&2 represented double digested hIL-6 recombinant plasmid with SalI and HindIII restricted enzymes from DF and DHF patient serum sample (data not shown for healthy controls).

Figure 1 1% Agarose TAE gel illustrating hIL-6 gene PCR amplified product (~640bp size).
Lane: 1 is showing the PCR amplified hIL-6 gene (~640bp) from normal healthy control.
Lane: 2-3 from DF and Lane: 4-5 from DHF patient’s serum sample. Lane M: 1000-bp DNA Marker.

Figure 2 Double digestion of hIL-6 recombinant plasmids isolated from white colonies and 1% agarose TAE gel is showing double digested hIL-6 fragment from TA vector. Lane M: 1000bp DNA Marker.
Lane 1&2: Double digested hIL-6 recombinant plasmid with SalI and HindIII restriction enzymes from DF and DHF patient serum samples.
Five of the positive clones confirmed through colony PCR and restriction analysis from DF and DHF patient’s serum were subjected to sequencing reaction with M13 universal sense and anti-sense primers. Complete sequences of the hIL-6 gene were obtained from DF patient’s serum as 639bp and 636bp from DHF patient’s serum with no ambiguity remaining. The derived sequences were subjected to an initial BLAST (Basic Local Alignment Search Tool) analysis. The hIL-6 sequences from DF patient’s serum shown complete nucleotide identity of 100% (data not shown) and DHF serum sample derived hIL-6 with the highest nucleotide identity of 99% with Homo sapiens IL-6 gene21 mentioned as an IL-6 original in Figure 3. Here we found one deletion of three bases GGG in our reported sequence, mentioned against IL-6 New in Figure 3. After BLAST analysis, the DHF serum derived hIL-6 sequence was reported to NCBI nucleotide database (GenBank: JQ250825.1). Further structural in silico analysis was performed only for DHF serum derived hIL-6 as it was consistently found to be altered from five DHF serum samples.
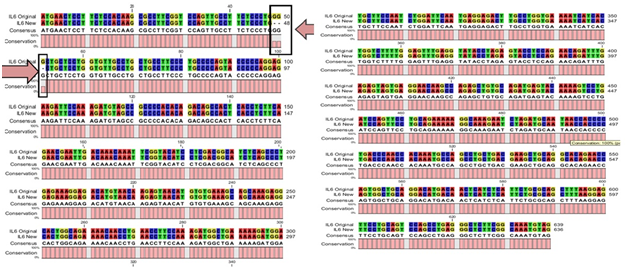
Figure 3 CLC workbench Alignment result of DHF serum derived hIL-6 nucleotide sequence (Accession No: JQ250825.1) to previously reported hIL-6 sequence (Accession No: NM_000600.3) used as a template.
In silico analysis
The predicted protein sequence for DHF serum derived hIL-6 (GenBank: AFF18412.1) was aligned to previously reported hIL-6 sequence22 using CLC workbench. The Glycine deletion at position 17 in protein sequence was depicted in Figure 4. Alignment result is illustrating Glycine deletion at position 17 in DHF serum derived hIL-6 protein sequence. To check the effect of Glycine deletion at position 17 in our hIL-6 protein sequence, a 2D model was predicted through online tool PSIPRED, presenting the number and location of its helices, coils and strands as shown in Figure 5A whereas, Figure 5B was the reference 2D model of hIL-6 protein sequence.21 Transmembrane protein topology of our predicted hIL-6 protein was guaranteed by depicting the location of its residues in cytoplasmic and extracellular regions. Signal peptide in our predicted secondary model of hIL-6 was located at position 1-26, while for reference secondary model was located at position 1-24. The N-terminal for both models was protruding out in an extracellular region with no re-entrant helices.
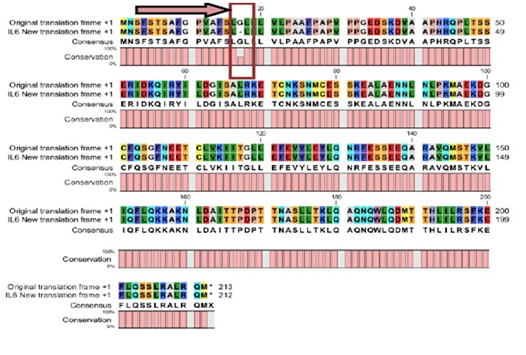
Figure 4 CLC workbench Alignment result of DHF serum derived hIL-6 protein sequence (GenBank: AFF18412.1) to previously reported hIL-6 protein sequence (Swiss-Prot: P05231.1) used as a template.
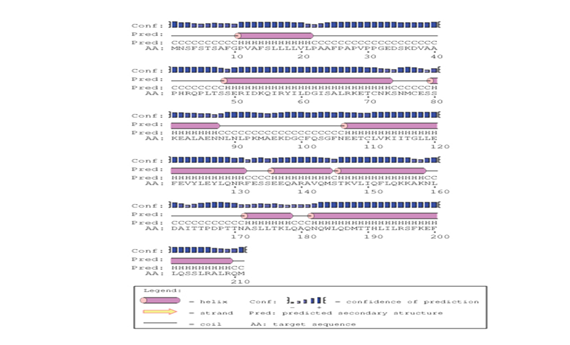
Figure 5A Predicted secondary structure of DHF serum derived hIL-6 (GenBank: AFF18412.1) Protein sequence through PSIPRED protein structure prediction server.
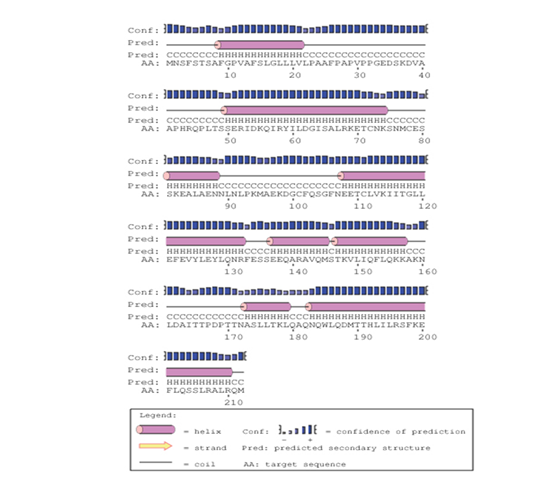
Figure 5B Predicted secondary structure of DHF serum derived hIL-6 (Swiss-Prot: P05231.1) Protein sequence through PSIPRED protein structure prediction server.
Moreover, predicted DHF serum derived hIL-6 protein sequence was submitted to NCBI conserved domain database to comprehend the deletion position through Live BLAST search. Similar domain architectures through CDSEARCH/cdd v3.15.0 excluded this deleted position 17 in our sequence from conserved region indicating this deletion event as an insignificant change (Figure 6). Here it became clear that query protein sequence comprised 212 amino acids in total and specific hits for conserved domain search in IL-6 Super family targeted the query sequence after the amino acid no.25. Therefore, the deleted position 17 in our hIL-6 protein sequence proved as to be insignificant been residing out of the conserved domain. A 3-D model of DHF serum derived hIL-6 was predicted through online tool i-TASSER shown in Figure 7A. This model was aligned to template hIL-6 protein model (PDB: 1P9M-B).23 The alignment showed query structure was in complete agreement with its homolog structure (Figure 7B). Our predicted 3-D model was subjected to PROSESS analysis to confirm its coordinates and to ensure similarity assessment with its homolog structures. The similarity was assessed through BLAST searches of the protein sequence through Protein Data Bank (Figure 8). PROSESS job information also depicted 56% helices, 44% coils, 9% turns and 0% Beta-strands in our predicted hIL-6 model. Later, the model was evaluated through ERRAT and PROCHECK tests for structural validity. Evaluation scores for our predicted model with PDB hit1 IL6-A as a template, were given in Table 1.24 The structures with expected values <10-7 were listed along with their resolution/Rfree values. The Root Mean Square Deviation (RMSD) between the input structure and the related structures were calculated and displayed. It was used to evaluate the structural homology between predicted and reported model. The structures significantly different from related structures consistent with their RMSD and sequence identity (IDEN) were flagged. The ranking of the model was based on function prediction score (Fh-score), which is a combined measurement scale for evaluating the global and local similarity between query and template protein. Fh-score values range in between;5 where a higher value indicated a better confidence in predicting the function using the template. IDEN was the percentage of sequence identity in the structurally aligned region. TM-score showed the measure of global structural similarity between query and template protein. Overall quality of the query structure agreed with most homologs. Quality factor generated by ERRAT test was 93.564 % rendering our predicted hIL-6 model with high resolution (Figure 9). Ramachandran plot produced as a result of PROCHECK analysis showed that 88.5% of the total residues were attending the most favored region (Figure 10).
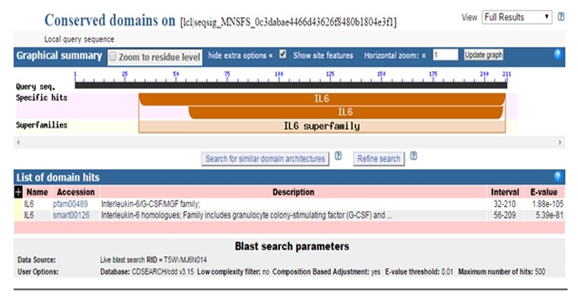
Figure 6 Description of conserved domain search result for predicted DHF serum derived hIL-6 protein sequence through NCBI Conserved Domain Database.
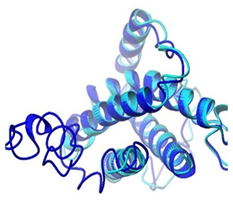
Figure 7B Alignment of predicted DHF serum derived hIL-6 model with previously reported hIL-6 structure (PDB ID: IP9M-B) Alignment result is illustrating query model (blue) in complete agreement with template model (magenta).

Figure 8 Similarity assessment results of predicted DHF serum derived hIL-6 structure with its structural homolog; (PDB ID: IP9M-B) through PROSESS job. The PROSESS job similarity assessment results indicated almost 100% identity of hIL-6 to its structural homolog.
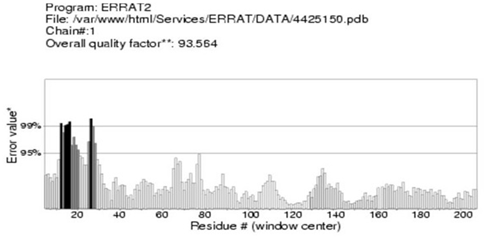
Figure 9 Evaluation of the DHF serum derived hIL-6 3-D model by ERRAT test. ERRAT score for evaluation of hIL-6 3-D model showing residues on X-axis and Error value on Y-axis: Overall quality factor calculated as 93.564% ensured high quality of the predicted model.

Figure 10 Evaluation of the DHF serum derived hIL-6 3-D model by PROCHECK test. Ramachandran plot is giving the location of residues in most favored, allowed and disallowed regions. The predicted hIL-6 model shown 88.5% of the total residues were attending the most favored region.
PDB HIT |
TM-Score |
IDEN |
RMSD |
Fh-Score |
Overall Quality Factor |
1IL6-A |
0.6937 |
0.97 |
2.15 |
3.20 |
93.564 |
Table 1 Validity estimation scores for predicted hIL-6 3-D protein model
Various studies have proved hIL-6 up-regulation during severe DV infection with fatal outcomes so the structural analysis of this gene is inevitable to correlate its function and behavior during DHF. Here we tried to analyze the hIL-6 gene structurally from DHF patient’s serum to inspect structural relevance or discrepancy to its homologs. The in silico analysis of over expressed hIL-6 gene showed a minor deletion in the sequence, keeping the structure aligned and identical to other homologs. To investigate the effect of one amino acid deletion at position 17 in our reported sequence, we predicted and structurally evaluated its protein 3-D model. Evaluation tests clearly depicted the model with best quality and identity to its homologs. We conclude from DHF serum driven over expressed hIL-6 gene in silico analysis that it is highly identical to normal healthy and DF homologs. This basic information for better understanding of advanced structural analysis of hIL-6 in future will not only be helpful for distinct intervention strategies to inhibit IL-6 overproduction during DHF but also help to design its antagonists for therapeutic purpose and in the broader spectrum to answer the questions about when and how to block IL-6 for DV-infected patient.
The project was Funded by the Natural Science Foundation of China (no. 31300603), the Program for Professors of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning (no. 2012-28), the National College Student Innovation Experiment Program (no. 14T10501), and the General Financial Grant from the China Postdoctoral Science Foundation (2015M571455) and we would like to express our gratitude to Dr. Najam Us Sahar Sadaf Zaidi and Dr. Rehan Zafar Paracha for their technical support and valuable suggestions during this research work.
There are no financial conflicts of interest.
None.

©2017 Kanwal, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.