MOJ
eISSN: 2373-4442


Short Communication Volume 1 Issue 4
Department of Microbiology and Immunology, Drexel University College of Medicine, USA
Correspondence: Brian Wigdahl, Department of Microbiology and Immunology, Institute for Molecular Medicine and Infectious Disease, Drexel University College of Medicine, 245 N. 15th Street, MS # 1013A, Philadelphia, PA 19102, Tel 215-762-7598, Fax 215-762-1955,+331-537-220-51
Received: August 24, 2014 | Published: October 17, 2014
Citation: Dampier W, Nonnemacher MR, Sullivan NT, Jacobson JM, Wigdahl B (2014) HIV Excision Utilizing CRISPR/Cas9 Technology: Attacking the Proviral Quasispecies in Reservoirs to Achieve a Cure. MOJ Immunol 1(4): 00022. DOI: 10.15406/moji.2014.01.00022
Recently several gene-editing technologies developed are being explored for their potential utility in providing new and unique treatments for HIV. One of these technologies is the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas)9 system. This system is being explored for its utility against host genes important to HIV infection, namely the HIV coreceptor CCR5, and for excision of the integrated genome from infected cells by targeting selected genes or genomic regions, especially the HIV-1 promoter or long terminal repeat (LTR). One of the major hurdles with the development of this technology for use in patients is defining the LTR sequence spectrum within the viral quasispecies present in the integrated virus and how that effects the number of guide RNAs (gRNAs) required to completely excise all proviral genomes. In this study, the Drexel Medicine CNS AIDS Research and Eradication Study (CARES) Cohort was utilized to demonstrate that1 the predominant sequence of the integrated proviral LTR within the PBMC compartment shows a decrease in the amount of variation per year regardless of the type of therapy;2 predominant HIV-1 LTR sequence undergoes continued genetic change with respect to the predominant genotype in these cells for at least 6 years while on effective suppressive ART;3 using next generation sequencing (NGS), to demonstrate that 4 of the 8 patient samples examined could have a complete gRNA regimen designed to target all known quasispecies; and4 length of HAART therapy may reduce the number of gRNA required to eradicate provirus as shown by NGS and gRNA design for longitudinal samples of patient A0017 in the CARES cohort. Overall, these studies demonstrate the feasibility of addressing at least one of the major technological challenges of CRISPR/Cas9-mediated HIV-1 proviral genome eradication involving the effective targeting of all viral quasispecies in a given patient sample.
Keywords: CRISPR/cas9, guide RNA, HIV, memory T cells, excision
CRISPR, clustered regularly interspaced short palindromic repeats; LTR: long terminal repeat; CARES, cns aids research and eradication study; vQS, viral quasispecies; HAART, highly active antiretroviral therapy; HDAC, histone deacetylase inhibitors; PKC, inhibitors and protein kinase C; ZFN, zinc finger nucleases; HERVs, human endogenous retroviruses: TALENs ,transcription activator-like effector nucleases; LTRs, long terminal repeats; ART, anti-retroviral therapy
According to the 2013 global report from the Joint United Nations Programme on HIV/AIDS (UNAIDS), 35.3 million people worldwide are infected with HIV-1 despite preventive and therapeutic measures.1 Furthermore, comorbidities and secondary factors such as drugs of abuse, non-compliance of drug therapy regimen, and evolving HIV viral quasispecies (vQS) complicate a cure. Patients who adhere to a highly active antiretroviral therapy (HAART) regimen typically maintain low or undetectable viral loads along with a near-normal CD4+ T-cell population. Initially it was hypothesized that patients remaining on HAART for extended periods would eventually be cured. However, not long after the success of HAART was realized, reservoirs of latently infected cells were discovered. These hidden cells produce minimal levels of viral protein and thus avoid both viral cytopathic effects and host immune clearance.2,3 The most prominent latently infected cell pool is thought to be the resting CD4+ memory T-cell compartment; however, depending on which end-organ one is referring to, such as the CNS, microglia and macrophages are likely the primary viral producer and reservoir.4,5 In addition, these reservoirs, particularly the CNS, are thought to be established shortly after the initial phase of infection. The dynamics relative to the establishment of viral latency and reservoirs has been previously reviewed in depth.3,4,6
The resting CD4+ memory T-cell population retains the capacity to produce infectious virus particles upon stimulation or cessation of HAART and thus are a major barrier to achieving an HIV cure2,3 and this remains the case even after prolonged periods of therapy. Current efforts to eradicate HIV-1 from the resting CD4+ memory T-cell population primarily focus on a “shock and kill” method where compounds are utilized to induce reactivation of virus from this cell population, through the use of compounds such as histone deacetylase inhibitors (HDAC) inhibitors and protein kinase C (PKC) inhibitors to allow the host immune response to recognize and target these infected cells. However several limitations have been realized in this type of therapeutic approach including: (i) there is a large fraction of non-functional genomes within this latent reservoir and therefore not all integrated provirus can produce replication competent virus,7 in essence leaving behind integrated HIV that may still be able to produce viral proteins or other components of the virus that could still have adverse effects, (ii) the overall number of CD4+ T cells reactivated from the resting CD4+ T-cell HIV-1 reservoir, as determined by viral outgrowth assays, has been quantified as being orders of magnitude smaller than the number of infected cells detectable by PCR-based assays, suggesting that not all cells within this reservoir are being reactivated,2,8 and (iii) the observation that the CTL immune response is not robust enough to eliminate infected cells following reactivation.9 Furthermore, the “shock and kill” method maybe less effective in cells of the monocytic lineage based on inherent differences in their physiology, particularly their innate resistant to cytopathic effects of HIV-1 and their terminally differentiated state that requires unique therapeutics, such as PI3K/Akt inhibitors and others.10,11 Given these observations, one of the current research objectives has been to find novel approaches to reduce the size of the latently-infected cell population that do not require activation of HIV gene expression or reactivation of virus production. To date, four gene editing techniques have been examined with respect to HIV eradication efforts and include zinc finger nucleases (ZFN), transcription activator-like effector nucleases (TALENs), piggyback,12 and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas)9 system.13
Recently the CRISPR/Cas9 system has been shown to have wide utility in genome engineering in yeast,14 Drosophila,15 human and mouse cell lines,16-19 and in zebrafish, mice, and C. elegans20,21 animal models. With respect to the use of this technology in the setting of HIV infection, several lines of research have been proposed including (i) disrupting HIV-1 entry coreceptors (CCR5, CXCR4) and proviral DNA-encoding viral proteins,13,22 CCR5 gene targeting ZFNs are in phase II clinical trials for HIV/AIDS treatment,23 (ii) engineering resistant cells by prior immunization with this type of therapeutic approach,24 (iii) prospects of selectively deleting HIV proviral DNA integrated into the host genome,13,22,25,26 (iv) removing the proviral HIV-1 genome from host cell DNA, by targeting its highly-conserved 5’ and 3’ long terminal repeats (LTRs),27,28 and (v) targeting specific cis-acting elements within the LTR.27 Recently these concepts and combined approaches were used to identify HIV-1 targets while excluding host off-target effects to accomplish the excision of an LTR fragment of the proviral DNA from latently-infected T cells and a 9.7kb fragment from microglial and promonocytic cells with no detected off-target effects and prevention of HIV-1 infection utilizing the Cas9 system.24 These studies have clearly shown great promise for other cell types using this system.
However, the CRISPR/Cas9 system is not without its complications. The system requires the design of a regimen of guide RNAs (gRNAs) that are identical to the 5’ and 3’ ends of the desired excision site. To reduce off-target effects, these cannot be homologous to the host genome; this is non-trivial due to HIV’s similarity to many human endogenous retroviruses (HERVs).29,30 Furthermore, HIV’s high mutability along with inter- and intra-patient variability make this a complicated problem. Additionally, targeting of the viral promoter or LTR involves transcription factor binding sites that are similar to the ones present in human promoters.
The gRNA design principles limit the breadth of possible sequence targets and it is unclear how many gRNAs can be delivered in a single regimen. Originally it was thought that gRNAs required an exact match of a 20 nucleotide primer followed by the NGG PAMmotif, but recent research by Hsu and coworkers31 has described a more complicated relationship. There is a non-linear relationship between the position of mismatches and the effect on subsequent binding, implying that predicting off-target effects will require more than a standard BLAST search approach. Furthermore, the requirement of a terminal PAM motif further limits the number of targetable positions in the HIV genome. As such, the combination of these factors along with the variability of the vQS indicates that there may be a subset of the HIV-infected population for which a complete regimen cannot be designed.
Some studies that have examined the resting CD4+ memory T-cell reservoir have shown that the viral quasispecies that can be induced from these cells starts to become more homeostatic the longer a patient is maintained on HAART.2 However, these studies did not consider the uninduced provirus. We have analyzed sequencing results from HIV-1-infected patients enrolled in the Drexel Medicine CNS AIDS Research and Eradication Study (CARES) Cohort32 which show that the predominant sequence of the LTR from integrated provirus within the PBMC compartment exhibits a decrease in the amount of variation per year regardless of type of therapy (Figure 1A) but that this virus still undergoes continued genetic change of the predominant genotype in these cells for at least 6 years while on effective suppressive ART with a constant median of 10-20 unique mutations per year throughout the entire LTR (Figure 1B). The amount of variation per year appears to be reaching a plateau around the six year mark but additional studies will be required to further document this apparent observation. Given these results, it is essential to use next-generation sequencing to determine all of the vQS present in a well-controlled patient’s reservoir. Using this approach will allow for the design of a gRNA regimen that will eliminate all vQS present with excision therapy. This would eliminate the need to understand if a given infected cell has a replication competent virus or not as in theory it would eliminate all HIV-1 targets present within a given cell population.
Utilizing Roche 454 next generation sequencing (NGS), we have performed NGS on genomic DNA isolated from PBMCs of 6 patients and 8 samples to gain an appreciation for the conservation of the HIV-1 LTR and number of gRNAs potentially needed per patient to target all known quasispecies. Due to the limitation of gRNA construction, a complete gRNA regimen could only be designed for 4 of the 8 patient samples (Figure 1C). However, of the ones that could be designed, no one patient visit sample needed more than 10gRNAs to target the entire quasispecies in a given patient. This shows that for a subset of patients, even those with viral loads above 50copies/mL, a regimen with less than 10 gRNAs can be devised; conversely, for another subset of patients, even if they have viral loads below 50 copies/mL, no regimen can completely excise their infection.
Of the NGS samples, two of the six patients were tested longitudinally 11 months apart. When looking at longitudinal samples from these patients, the amount of variation in the LTR becomes more conserved (however not lacking of genetic variation) across the entire sequence (data not shown). One of these patients, patient A0017, presents an interesting case. At the intake visit, at which point they had been on HAART for six years, patient A0017 could not have been completely treated with CRISP/Cas9 excision therapy. Over the course of another year of HAART therapy, it was possible to design an excision regimen of less than 10gRNAs at various sites to be able to cover all the known quasispecies from the PBMC population (Figure 1D). This indicates that the vQS is a moving target and the therapeutic window will be limited for a given set of gRNAs even with the most effective therapies currently available. This also indicates the need for sequencing at a genome level to allow more advanced selection of gRNAs from multiple regions rather than shorter reads that 454 or Illumina sequencing strategies will provide. Taken together, these results have suggested that using NGS techniques can result in not only identifying all quasispecies present in cell populations but also that the number of gRNAs needed will be low enough to potentially be packaged into delivery systems such as any of a number viral vectoring strategies. As demonstrated, the challenge of targeting all of the integrated viral LTRs will be complex but certainly feasible.
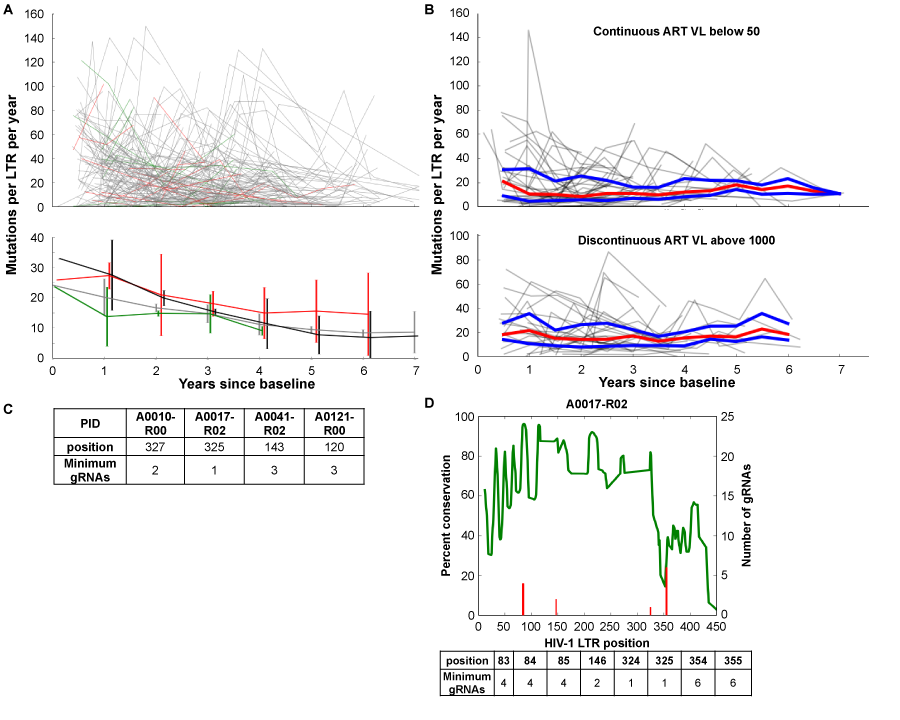
Figure 1 HIV LTR genetic variation and gRNA design in well-controlled patients. Consecutive visits were compared by individually aligning all sequences from each patient using the MUSCLE alignment tool33 and calculating the number of variations between consecutive visits. The number of nucleotide changes per 100bp was plotted against the time since the baseline visit to determine the rate of accumulated variations.
A: Line segments from each patient were generated from the longitudinal variation based on the anti-retroviral therapy (ART) status between consecutive visits using the following coding scheme: green line segments indicate a longitudinal visit in which the patient was naive to ART (21 patients), red line segments indicate off/non-adherent ART visits (39 patients), grey line segments indicate on/adherent ART visits (168 patients), and black line segments on/adherent ART (54 patients) with viral loads always below 100 copies per ml. The top panel depicts all longitudinal samples per patient. The bottom panel shows median and standard deviation for each group at each year.
B: LTRs from 45 patients on/adherent ART and 31 patients with discontinuous ART for at least three consecutive visits were analyzed as in A. The trajectory of each patient is shown in grey with the median in red and the upper and lower quartiles in blue.
C: Utilizing Roche 454 next generation sequencing (NGS), NGS on genomic DNA isolated from PBMCs of 6 patients and 8 samples was performed on a 4.4kb fragment of the HIV genome, as previously described.32 The number of gRNAs and target position for each patient sequenced was determined by aligning the short-read sequences to the HXB2 genome using the BWA aligner34 and a local implementation of the algorithm used by the CRISPR design tool.24 23-mer sliding windows were constructed by extracting all completely overlapping reads and checked for a PAM sequence; all windows with less than 50 overlapping reads were excluded. The minimal number of gRNAs required to cleave each targetable window was calculated by testing all possible gRNAs.
D:NGS reads from patient 17 visit 3 were mapped to HXB2 using the BWA aligner34 as described above and examined for percent conservation (green line) and number of gRNAs necessary for excision of all known quasispecies (red line) at every position of the LTR. A table of the position and number is provided below the graph.
Along with the challenge of designing the gRNAs necessary for the excision of all integrated virus, the other major challenge will be delivery of the gRNAs. One way that has been hypothesized is through the use of lentiviral vectors. This type of delivery system would allow the infection of similar cell types that HIV naturally infects, potentially limiting the delivery of the therapy to uninfected off target cells. However, the efficiency of using these to deliver to latent cells may be complicated by both the infectability of these types of cells as they are known to have a decreased ability due to the inactivity of the cell as well as cells, which have been infected by HIV having a more resistant phenotype to secondary HIV infection. Another limitation of this type of system will undoubtedly be how many gRNAs can be packaged into one vector to ensure that when the lentivirus vector system does infect a cell that all the potential gRNAs needed will be present. What the limit will be has yet to be defined. Finally with delivery systems like lentivrus vectors even if the above mentioned hurdles can be surpassed, delivery of the gRNAs to tissue resident cells which can have very low numbers and require migration across vascular barriers will be an additional level of challenge.
Given these challenges, immediate studies are needed to explore these issues. With respect to the viral genotype the first question will center on the genetic makeup of the vQS retained in reservoir cells like the CD4 memory T-cell population and whether these viruses are similar to or significantly different than the vQS retained in cells of the monocyte-macrophage lineage and then to define these differences with respect to viral eradication involving LTR targeting? Deep sequencing studies in well-defined patient populations will need to be performed using long fragment PCR techniques for proper gRNA design. Furthermore, viral genotype compartmentalization in other tissues may complicate the picture with the introduction of unique viral genotypes as compared to the peripheral blood. However, deep sequencing studies may find that there are viral regions of high conservation even across multiple tissues.
Second, what are the viral dynamics in these cells? How long must a patient be on ART to drive virus selection in the reservoir to a level where the number of quasispecies present is low enough to limit the gRNAs needed to eradicate the retention of viral proviral DNA in these tissues? Sequencing of patients, especially at cell subpopulation levels, to have a complete understanding of viral dynamics in well-controlled patients will be required. This challenge may be addressed by advances in the delivery of the CRISPR/Cas9 system allowing larger cassettes of gRNAs to overcome the variability observed even after prolonged therapy.
Finally, ex vivo proof-of-concept studies beginning with the memory T-cell population will be required to determine if virus can be eradicated from HIV-1-infected patient derived cells in a well controlled experimental environment within single cell T-cell populations cultured in vitro from HIV-1-infected patients.
Overall, while this type of HIV-1 eradication technology has many challenges, the prospects of this technology delivering a curative therapy remain very exciting. This is highlighted by the fact that the literature in this area is expanding at a rapid rate as well as by the fact that as mentioned earlier phase II clinical trials for an HIV/AIDS treatment using this technology with CCR5 gene targeting is already underway.23 Even with this observation, it is reasonable to expect that this type of therapy is still at least 5-10 years away from being able to be considered as an effective therapy in humans. However, with the ever changing rate of experimental advance perhaps the timeline will be more quickly than expected. This may depend on difficulties encountered in delivery technologies to facilitate studies designed to attack HIV-1 reservoirs beyond the peripheral blood and regional lymph nodes. At this point, the challenges still outweigh the successes to date in the early stages of development; however, this is usually a true statement for any new therapeutic approach. Clearly, the excision strategy represents the only experimental therapeutic strategic that may achieve complete elimination of chromosomal based defective and activation competent proviruses. In the not to distant past, this seemed to be an unachievable objective; this no longer seems to be the case.
These studies were funded in part by the Public Health Service, National Institutes of Health, through grants from the National Institute of Neurological Disorders and Stroke, NS32092 and NS46263, the National Institute of Drug Abuse, DA19807 (Dr. Brian Wigdahl, Principal Investigator), National Institute of Mental Health Comprehensive NeuroAIDS Core Center (CNAC), P30 MH-092177 (KamelKhalili, PI; Brian Wigdahl, PI of the Drexel subcontract), and under the Ruth L. Kirschstein National Research Service Award 5T32MH079785 (Jay Rappaport, PI, Brian Wigdahl, PI of the Drexel subcontract). The contents of the paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. Drs. Michael Nonnemacher and Will Dampier were also supported by faculty development funds provided by the Department of Microbiology and Immunology and the Institute for Molecular Medicine and Infectious Disease.
There is no conflict of interest.
None.

©2014 Dampier, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.