MOJ
eISSN: 2573-2919


Opinion Volume 3 Issue 1
Faculty of Environmental Protection, Forestry and Forest Engineering Department, University of Oradea, Romania
Correspondence: Ecaterina Fodor, Faculty of Environmental Protection, Forestry and Forest Engineering Department, University of Oradea, Street Gen, Magheru, no.26, Oradea, Romania
Received: October 24, 2017 | Published: January 31, 2018
Citation: Fodor E, Network science and forest species interactions. MOJ Eco Environ Sci. 2018;3(1):33-36. DOI: 10.15406/mojes.2018.03.00063
Network science, a relatively young science discipline triggered important developments in ecology leading to the emergence of a new domain, network ecology. Important contributions to the study of species interactions were made using the approach of bipartite networks depicting pair wise interactions between two sets of species. At the moment, a new research trend based on the interlinked networks pervades network ecology. A heuristic network is proposed showing the possible links among different guilds of organisms directly or indirectly linked to focal forest species, the trees. The network is a possible roadmap for future research directions aiming forest ecosystems.
Keywords: Network science; Network ecology; Network of forest guilds; Roadmap
It is already common knowledge that ecology benefits from advances in various fields of mathematics. Network theory1 introduced new opportunities in exploring species interactions incorporating trophic, mutualistic, facilitative, antagonistic relationships, by targeting superior level of integration compared to pair wise approach of classical ecology. Interactions are key elements of structural community properties like biodiversity,2 community assembly rules, ecological niches, energy flow through the trophic webs, nutrient cycles to name few most important. The hidden properties of interacting species networks are revealed through metrics and analytic tools produced by network science. Other important contributions of network theory to ecological topics consist in the analysis of habitat structures at landscape and meta-ecosystem levels,3,4 disease spread, biotic invasions and, at temporal scale, co-evolution of mutualist /parasites and their hosts5,6 and conservation ecology.7 At the moment, we witness the emergence of a new discipline within the large domain of ecology, namely network ecology.8 Networks are flexible conceptual models providing a systematic way to approach complexity and addressing practically any type of system with interlinked elements. Network science studies networks and is grounded in graph theory being an empirically driven domain with elements borrowed from engineering (control and information theory) statistical physics and social science1 also a fast growing discipline. At the moment, we witness the emergence of a new discipline within the large domain of ecology, namely network ecology.8 One special category is represented by ecological networks that are non-random, complex and heterogeneous structures, with small world and scale free topology9,10 and with a well-defined architecture.11 However, compared to technological or social networks, metabolic pathways or protein-gene interactions,12,13 they are relatively small in terms of number of nodes10 and number of interactions.
Basically, networks are composed of nodes corresponding for most cases in ecological networks to species, and arc or edges connecting nodes corresponding to undirected or directed links or interactions. Ecological networks are depicting unipartite trophic webs and bipartite networks (two sets of nodes linked between but not within sets) depicting antagonistic or mutualistic interactions. The topology of networks involving different groups of organisms appears to be highly specific for the type of interactions and type of organisms involved. This specificity is proved by the existence of common general determinants of networks’ architecture, estimated through keystone metrics: connectivity, node degree distribution, nestedness and modularity that shape differently trophic, antagonistic and mutualistic networks. Other metrics are devised to describe network architecture in most studies on bipartite networks are less frequently used such as interaction strength and asymmetry, web asymmetry, togetherness, vulnerability, niche overlap, C-score, in fact, metrics previously used in community ecology and transferred to network analysis.14 Connectance is the first network topological property to be studied in ecological networks. Since the seminal papers of Margalef & Gutierez,15 and Jordano,16 connectance became the basic metric calculated for any studied network expressing to the number of realized links in a network.17 In time, the study of network topology became increasingly sophisticated,18 centered on the analysis of the distribution of links, the clustering patterns. Node degree distribution is a summary of complex networks’ topology.19 It is a basic network descriptor which provides the probability that a random selected node in a network has degree k.1 Biological networks (including ecological networks) present a specific pattern of the interactions or links among nodes fitting exponential, power law (or scale free) and truncated power law distributions.5 Nestedness, applied mostly to bipartite networks, a metric coined from biogeography and community ecology20 occurs when specialist species tend to interact with proper subsets of the species that interact with more generalist ones, considered by many researchers but not all, as an emergent network property.21 Nested subset structure was described among others, in host-parasite systems22 facilitation plant networks,23 ant–plant mutualistic networks,24,25 commensalism relationship between trees’ epiphytes26 and mycorrhizal systems.27,13,28 Nestedness has important consequences; it makes the community more robust against extinction29 stabilizes communities and enhance persistence.30 It was stated that nestedness varies according to antagonistic and mutualistic interactions11 a conclusion questioned by other authors.Modularity is considered an important network property underlying the maintenance of biodiversity,31 also a universal property of biological systems.32 Modularity occurs when groups of species interact more within own groups than among groups, a hallmark of systems heterogeneity leading to the specific small world topology.33,16,18 Since any ecological network is an opened system to which new nodes can be attached, modularity is a consequence of a more general property, the preferential attachment. The growth of a network shows that new nodes attach to the most connected in9 plain words expressed as rich get richer. The modular structure of complex networks plays a critical role in their function34 meaning that different groups of nodes play different roles with some degree of independence caused by multiple factors35 also community search and the identification of functional clusters give key insight on dynamics of properties of ecological communities.36 Modularity is linked to many important community properties as for instance in the evolutionary mechanisms involved in the assembly of host–pathogen systems37 and increase stability providing a mechanism by which complexity of the system can persist.38,18
Excepting food webs, most of the investigated ecological, highly resolved networks are bipartite depicting the properties of pair wise combinations of antagonistic or mutualistic nature. However, bipartite networks are building blocks for more complex ecological webs39 and are opened permitting the addition of new species (nodes) as new information is acquired. The question whether bipartite networks reflected community properties leading to stability looking exclusively at the properties of a single interaction type40 opened a new path in network studies; merging networks and analyzing pair wise interactions in complex. Those interlinked networks present novel topologies to be investigated. If considering only fungi as a key group in any terrestrial ecosystem, during the last years, papers on fungi and their complex interactions with other organisms (plants or animals) increased in number in a rapid pace. The binary interactions (a functional group of fungi such as ectomycorrhizal or pathogenic) were extended to more inclusive interlinked sub-networks relating fungal diverse soil biota to plants.41,42 Trees accumulate a great diversity of direct and indirect interactions, guilds with various levels of specialization (galling invertebrates and mining insects are by far the most specialized partners) due to their complex architecture and long life span.43 They establish also dominant populations in specific complex terrestrial ecosystems, the forests. For illustration purpose, I propose a heuristic, unipartite, un-directed network linking different guilds centered on forest trees, a map of interactions to be merged. Every two connected guilds correspond to a possibly species level resolved bipartite network. Linking all these pair wise combinations would give a comprehensive image of tree dependent biodiversity in terms of species and links. Building the guilds’ network is a strictly heuristic approach and is based on documented interactions.
Merging different networks depicting interactions of different levels of intimacy sensu Thompson,31 generates indirect links among different guilds, one can speculate that latent network properties could emerge and give new and unexpected explanations on how communities assemble and how diversity functions, and also widely used network metrics would vary in a meaningful way. The links among guilds are documented using information from published articles and books. Mutualist interactions, with different levels of intimacy, establish direct and indirect links with trees; from pollinators and facilitators such as nurse plants and seed dispersers to phytophagous insects’ mutualists and mycorrhizal helper. Antagonistic guilds contain directly associated organisms to trees as pathogens and insects attacking different tree organs, but also their antagonistic or mutualistic associates such as hyper parasitic fungi and insects (for details see Figure 1 with explanations). Modularity and connectance were assessed in order to have a rough estimate of network’s architecture: low connectance of 0.04 and relatively high modularity of 0.53 (10 modules identified) characterizes the proposed network. One important aspect is the presence of numerous guilds of parasites and pathogens, formerly neglected groups in the functional structure of the biocoenoses, reconsidered as a common consumer-resource assembly,44 and attached to every free living or mutualistic guild, important contributors to overall biodiversity.45–48
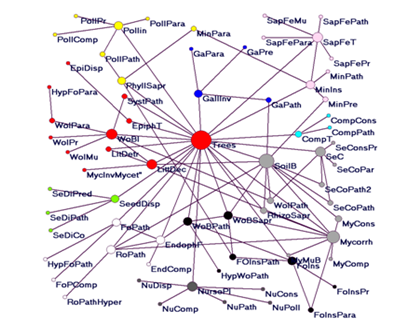
Figure 1 Network mapping the possible pair wise combinations of interspecific interactions (antagonistic and mutualistic). Considering trees as focal organisms, nodes correspond to distinct guilds encompassing antagonistic and mutualist interactions, above and belowground. The following abbreviations correspond to direct interactions between trees and distinct functional groups of organisms, Mycorrh, Mycorrhizal Mutualists; Nurse, Seedlings’ Facilitators Represented by Shrubs or other Tree Species; Pollin, pollinators; CompT, tree competitors; EpiphT, epiphytic plants and lichens including parasitic and semi-parasitic species; EndphF, endophytic fungi functioning as commensal organisms or mutualists in mostly all tree organs; LittDec, tree foliar litter decomposers; RhizoSapr, saprotrophic species from the rhizosphere of trees; SeC, seed and fruit consumers; SoilB, soil biota encompassing invertebrates, Bacteria, protists and fungi interacting with trees; PhyllSapr, phylloplane saprophytic bacteria and fungi; seedisp, seed and fruit dispersers (Facilitators); RoPath, root pathogens; FoPath, foliar pathogens; Sometimes opportunistic endophytes; WoBPath, wood and bark pathogens and parasites; WoBSapr, wood ad bark saprophytes; SystPath, systemic and wilting pathogens; FoIns, foliivorous insects; GallInv, galling invertebrates minins, mining insects; SapFeT, sap feeding insects on trees; WoBI, eood and bark boring insects. indirect links going from trees to interacting partners of the primary functional groups; MyMuB, mutualistic bacteria of mycobionts; MyComp, competitors of mycorrhizal partners; MyCons, consumers of mycorrhizal fungi, either belowground Mycelia or above ground carpophores; SeCoPr, predators of seed and fruit consumers; SeCPath, pathogens of seed and fruit consumers (Targeting Insects); SeCoPath2, pathogens of seed and fruit consumers (targeting vertebrates); SeCoPara, parasitoids of insect seed and fruit consumers; NuComp, competitors of nurse plants such as herbaceous plants; NuCons, consumers of nurse plants, for instance defoliating insects or large mammal browsers; NuDisp, nurse plant dispersers, including frugivorous birds and small mammals; NuPoll, pollinators of nurse plants; NuPath, nurse plant pathogens; Comp Cons, consumers of trees’ competitors; CompPath, pathogens of trees’ competitors; GaPara, parasitoids of the galling insects; GaPath, pathogens of the galling invertebrates; GaPre, predators of galling invertebrates; WoIPr, predators of wood and bark insects; WoIPara, parasitoids of wood and bark insects; WoIPath, entomopathogens of wood and bark insects; WoIMu, mutualists of wood and bark insects, such as protists, yeast or other fungal species; SeDiPred, predators of seed dispersers; SeDiPath, pathogens of seed dispersers; SeDiCo, competitors of seed dispersers; FoInsPr, predators of foliivorous insects; FoInsPath, entomopathogens of foliivorous insects; FoiInaPara, parasitoids of foliivorous insects; LitDetr, litter detritivorous invertebrates; FoPComp, competitors of foliar pathogens; EpiDisp, dispersers of epiphytes; MinPre, predators of mining invertebrates; MinPara, parasitoids of mining invertebrates; MinPath, pathogens of mining invertebrates; SapFePr, predators of sap feeding insects; SapFePara, parasitoids of sap feeding insects; SapFePath, entomopathogens of sap feeding insects; HypFoPara, hyper parasites of foliivous insect’s parasitoids; HypFoPath, hyper parasites of foliar pathogens; MycInvMycet, mycophagous invertebrates and mycetozoans feeding on litter decomposers; Ropath Hyper, hyper parasites of root pathogens; sapFeMu, mutualists of sap feeding insects; HypWoPath, hyper parasites of wood and bark pathogens. the size of the vertices is proportional to the number of links. 10 Modules are depicted in different colors attached to corresponding groups of nodes. Louvain algorithm was used as it is provided by the software Pajek.46,47 The graph was generated in Pajek 32 5.01.
The graphical depiction of the guilds network shows that most diverse modules in terms of species and links are determined by trees. The next important modules or hubs are generated by extremely diverse and taxonomically heterogeneous soil biota and mycorrhizal partners, as mycorrhizal fungi interact with trees but also with soil biota, rhizosphere saprotrophs, root pathogens. The network can be interpreted as a roadmap in future forest ecosystem and forest community studies and give a rough approximation of the extent of connected biodiversity centered on trees. Since the famous interrogation in the title of Hutchinson’s paper “Homage to Santa Rosalia or why there are so many kinds of animals?” [1959) mechanisms and causes of biodiversity stirred constantly not only ecology but many other domains. Apparently, we are closer to at least a partial answer if we approach biodiversity from the angle of species interactions using the principles and tools of network science: a major contribution to the global science and the driving force of a new discipline, network ecology.
None.
None.
None.

©2018 Fodor. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.