MOJ
eISSN: 2641-9297


Research Article Volume 1 Issue 1
1Institute of Genome Research, Vietnam Academy of Science and Technology, Vietnam
2Centre of Aquaculture Biotechnology, Research Institute for Aquaculture No 1, Vietnam
Correspondence: Thuy Thi Bich Vo, Institute of Genome Research, Vietnam Academy of Science and Technology, Hanoi, Vietnam, Tel +84-24-32191174
Received: November 26, 2017 | Published: January 10, 2018
Citation: Bich Vo TT, Nguyen HD, Nguyen TTH, et al. Identification and analysis of snps in population of Vietnamese catfish (pangasianodon hypophthalmus), using next generation sequencing and snp validation. MOJ Curr Res & Rev. 2018;1(1):12-19. DOI: 10.15406/mojcrr.2018.01.00003
Tra catfish (Pangasianodon hypophthalmus) is a commercially important aquaculture species in the Mekong Delta, Vietnam; accounting for approximately 60% of Vietnam freshwater fish production. However, the food trade globalization and the growing demand for selected varieties of food have led to the intensification of commercial fraud, especially in the form of substitution species and mixing with cheaper taxa. DNA bar-coding methods have shown a potential well proven molecular approach to assess the authenticity of food items. In this study, we utilized high throughput NextSeq500 sequencing to capture and identified SNP markers on 400 wild and farming catfish individuals from 8 Vietnamese provinces (e.g., An Giang, Dong Thap, Can Tho, Tien Giang, Ben Tre, Hau Giang, Vinh Long, and Ho Chi Minh City) and 10 international locations of Cambodia, Thailand and Bangladesh. Stringent filtering of SNP-calling parameters resulted in 11,009 SNP loci represented across all 4 countries. We utilized the VCF tools to validate two panels of SNPs selected from the NGS data. Selection criteria included SNPs shared between catfish's Vietnam and international populations, and SNPs specific to each population. A total of 780 and 809 SNPs were filtered in Vietnamese and international catfishes, respectively, the final 12 special SNPs were detected. SNP validation by using Sanger sequencing and PCR showed a result of 2 per 12 SNPs was confirmed successfully in 300 individuals from Vietnamese catfishes. The first times our results provide genotype informative marker loci in Vietnamese Tra catfish in order to approach molecular traceability in aquaculture species with minimal previous genetic information.
Keywords: vietnamese tra catfish, next generation sequencing (NGS), single nucleotide polymorphism (SNP)
AS, allele specific; NGS, next generation sequencing; SNP, single nucleotide polymorphism; PCR, polymerase chain reaction
The Vietnamese striped catfish (locally name as Tra catfish) has been cultured in the Mekong Delta, from the 1960s of the last century.1 Average output of Tra catfish reaches 1.5 million tons per year, and has profited 1.6 billion USD in export business in 2016, increasing about 7% compared to 2015. Vietnamese catfish products have exported to 140 markets in the world, and the United States continued to be a major market for Vietnam's pangasius exports, accounting for about 20% of total exports.2 Vietnam has become the third largest producer of fisheries products in the world.3 However, in recent years, Vietnamese Tra catfish export industry has been facing many challenges such as illegal fishing, no declaration of origin, failing to comply with regulations and technical standards on food safety conditions and commercial fraud in fisheries. These stories have a huge impact on the economic and sometimes even public health implications. Therefore, the development of an efficient seafood traceability framework is crucial for the management of sustainable fisheries and the monitoring of potential substitution fraud across the food chains. There have been relatively few genetic studies of Tra catfish and information on genome resources in this species also limited.4-6 Next generation sequencing (NGS) technologies have opened many opportunities to develop molecular resources in biological and economic interest. In addition, genomic datasets represent may be mined in order to indentify suitable candidates for SNP marker development. SNPs potentially can be utilized for developing aquaculture lines7 or seafood traceability.8-10 Recently, over one hundred studies of whole genome sequences, transcriptome analyses, gene expression patterns, ESTs, and SNP identification for aquaculture species using NGS have been conducted to date, in particular for Atlantic salmon, catfish, rainbow trout, striped bass, oyster, zebra fish and medaka.11 The available genetic tools for traceability are SNPs, representing sites in the genome with minute mutations (novel genetic differences) in the DNA sequence. They are very abundant and widespread. Analyses of SNPs reach hitherto levels of population identification, rendering them optimal tools in fundamental biology, conservation and traceability.12 In the current study, the first times provide a genotype informative in Vietnamese striped catfish P. hypophthalmus using the NextSeq500 platform, Sanger sequencing and PCR.
Fish sources and sampling
All procedures involving the handling and collection fish samples during this study were approved by the Research Institute for Aquaculture No.1, Vietnam prior to initiation of the project. A total of 400 individuals catfishes, with 24 locations (i.e.,14 locations in 8 provinces of Vietnam and 10 locations in 3 countries as Cambodia, Thailand and Bangladesh- hereafter referred to as international samples) were used for this study. These locations were from different geographic areas, which possess different production traits such as growth rate, disease resistance and feed conversion efficiency. The original fish samples were collected from the Hau Giang River, Tien Giang River, the local fish farms (in Vietnam), Mekong river (the region of Cambodia, Thailand, and Bangladesh), and other local fish farms of 3 international countries (i.e., Strung treng, Kratie, and Tonle Sap of Cambodia; Loie, Nakhon Phanom, and Ubon Ratchathani of Thailand; Jessore, Brahmanbaria, Jamalpur, and Badal of Bangladesh). Catfish fins (approximately 300-500 mg) were sampled from different locations, placed in eppendorf micro tubes with absolute ethanol and maintained in a freezer at -20°C.
DNA extraction and sequencing
Total DNA was isolated using the GeneJET Genomic DNA Purification Kit (Thermo Fisher Scientific, CA, USA) following the manufacturer’s protocol. Equal amounts of DNA (100mg) from each individual were pooled for sequencing, one pool for each location. Sequencing was conducted at Institute of Genome Research (Hanoi, Vietnam). Genomic libraries were prepared with the TruSeq® DNA PCR-Free (Illumina, San Diego, CA) with 5mg of genomic DNA for all samples, according to the manufacturer’s instructions. For each pool sample, the prepared DNA library was sequenced on one lane of the Illumina NextSeq 500 platform for 100bp paired-end reads.
Reference mapping
Sequence mapping was performed using CLC Genomics Workbench (version 4.0.2; CLC bio, Aarhus, Denmark). Before mapping, adaptor sequences, ambiguous nucleotides (N’s), extreme short reads (<30bp) and low quality sequences (Quality score <20) were trimmed using CLC Genomics Workbench. After that, all trimmed reads were then aligned with the Vietnamese catfish genome reference (unpublished data). The mapping settings were: mismatch cost of 2, deletion cost of 3 and insertion cost of 3. The alignment had more than 95% similarity with the reference sequence across 90% of their length was accepted. The mapping output was export as BAM format13 for further analysis.
SNP identification and filtering
SNPs were identified from the alignment sequences using the SAM tools (version 0.1.18)13 and PoPoolation214 with the lowest criteria setting to obtain all potential SNPs. Two factors that are important for excluding false SNPs caused by sequencing errors were set: minimum read depth and maximum read depth. SNPs were defined as specific SNPs if the SNP polymorphisms were found in more than 95% organisms from only one location and less than 5% organisms from the other location. SNPs that were polymorphic in all strains were considered as common SNPs.
Polymerase chain reaction (PCR) amplification using primers for non-informative markers and Sanger sequencing
Genomic DNA was amplified by PCR using the selected primers (Table 1) in a 25μL PCR reaction consist of 12.5μL GoTaq Green Master Mix (Thermo Fisher Scientific, CA, USA), 1.0μL (10 ng) DNA template, 0.5μL of each primer (10 ppm), and 10.5μL sterile deionized water. Amplification was done in a Veriti Thermal Cycler (Applied Biosystems, Forster City, CA, USA) programmed for an initial denaturation at 94°C for 5min, followed by 25 cycles of 94°C for 30s, 45°C to 65°C based on the primer sequences for 30s, and 72°C for 50 min, with a final extension at 72°C for 10 min. The PCR products were visualized by standard 2 % agarose gel electrophoresis and purified by GenJet PCR Purification Kit (Thermo Fisher Scientific, CA, USA). A 10μL sequencing reaction volume was prepared using 2μL of the cleaned PCR product, 0.5μL (10 μM) of either the forward or reverse primer, 1μL BigDye terminator v3.1 (Applied Biosystems), and 1 μl Big Dye Sequencing (Genetix). The sequencing PCR consisted of 25 cycles of 10 seconds at 96°C, 5 sec at 50°C, and 4 min at 60°C. The sequencing reaction product was cleaned through sephadex columns before adding 20μl of Hi-Di formamide (Applied Biosystems) and then denaturation at 98°C for 2 min. The samples were analyzed on an ABI 3500 Genetic Analyzer (Applied Biosystems) with a 36 cm capillary and POP-6 polymer and using the default settings. The resulting parental sequences were visually screened for SNPs in Sequencher 4.2 software (GeneCode).
No |
Sequencing (5’-3’) |
Size of PCR product (bp) |
Annealing temperature (T°C) |
|||||
SNP 1 |
Forward |
AGAGCATAAGCAAATCGCTGC |
531 |
59°C |
||||
Reverse |
TTCCACTTGGGGCTTTGTTG |
|||||||
SNP 2 |
Forward |
TGGTATTCGGAGGAAAGATGGC |
654 |
57°C |
||||
Reverse |
TTGGACCACTAGGAGGAGACA |
|||||||
SNP 3 |
Forward |
GCTGTTCGGTTCAGAAGCTG |
617 |
59°C |
||||
Reverse |
AGCACAGGTGAAGGGTTGTC |
|||||||
SNP 4 |
Forward |
CACCCTGTGGACACGAACA |
516 |
59°C |
||||
Reverse |
AGCTGACTTCTCTAGCGGAC |
|||||||
SNP 5 |
Forward |
CCAGTGGTCATCTGGGGAAAG |
580 |
63°C |
||||
Reverse |
CGTCACTCACACACCACAAG |
|||||||
SNP 6 |
Forward |
AGTGCCATTGCTGTGGTAAC |
626 |
57°C |
||||
Reverse |
CACAGTCATGGCGTTGGATG |
|||||||
SNP 7 |
Forward |
GGCCGTGCTGTTATGCGAAA |
504 |
65°C |
||||
Reverse |
CATTCAGCAGCAATCAGGAGC |
|||||||
SNP 8 |
Forward |
CGGGTGTCCACCATGCTTTA |
559 |
63°C |
||||
Reverse |
GTAGCCGCACCATTCTCAGT |
|||||||
SNP 9 |
Forward |
CCCCAGGTTTGACATTGCAC |
650 |
63°C |
||||
Reverse |
CATCAGCGCCTGATCTCACT |
|||||||
SNP 10 |
Forward |
TCTGGGTGGACAAAGGTAGG |
528 |
63°C |
||||
Reverse |
CATGCACAACAGCTCACTGC |
|||||||
SNP 11 |
Forward |
GGCCTACGACGATGGTTACA |
532 |
63°C |
||||
Reverse |
CAGTTTAGGGAAGGCCCACA |
|||||||
SNP 12 |
Forward |
TAGTCAGACGCTCCAAACCG |
518 |
65°C |
||||
Reverse |
GAACCGAGCGTAGAAGTCGT |
|||||||
Table 1 Primer for PCR amplification
SNP validation with allele-specific (AS) markers by Nested-PCR amplification
Two SNP markers were selected from the mergered of sequencing results and reference sequencing. The AS primers were designed by a high throughput web tool for picking PCR and sequencing primers as BatchPrimer3 v1.0, containing SNP5- 45:G>A (45 indicates the position of the selective nucleotide) with the primer sequence, forward 5'-GCGAGGAGATTCACTCACAA-3', reverse 5’-GAGTGGCTATGAGTTGGCTC-3' (fragment size: 114 bp), and SNP9- 66:T>G with the primer forward 5'-AGCAAGATACAGCATTACCG-3', reverse 5’- GACTCACCTGCATGCGTAAT-3’ (fragment size: 145 bp). The first PCR amplified a region of SNP 5 and 9 using the designed primers in (Table 2). This step was performed to isolate regions of interest containing the relevant SNP 5 and 9 polymorphisms that were later used for the second allele-specific PCR to avoid amplification of similar sequences in the catfish genome that may be located outside the gene. After a successful the first PCR, 2.0 μL of diluted PCR product were used as a template for the detection of mutant in the second PCR. The second PCR was carried out in a reaction mixture and PCR condition with the modifications; higher annealing temperature (3-5°C increase) and reduced MgCl2 concentration (1.5 mM), which further increased the specificity of amplification. Amplicons were detected by electrophoresis in 2%-agarose gels and 1× TBE at 130 V for 90 minutes. SNP validation was confirmed from genomic DNA of all 400 individuals of the mapping population (including 300 and 100 individuals of Vietnamese and international catfishes, respectively).
No |
Sequencing of SNPs (Trimmed Seq) |
SNP position |
SNPs |
Call rate |
Location |
SNP 1 |
TGCAGCTGTAGCTGAATTGTAAAGTGTGCCTGGAGCCCTT |
28 |
C>T |
1 |
Vietnam |
CCTTCCAATTAAATGCAATGCATG |
|||||
SNP 2 |
TGCAGTCATGATGTGTTTCTTAAACTGGATGCATAGTTACT |
17 |
T>C |
0.989247 |
Vietnam |
GAGCAACACACACACACATACACACACA |
|||||
SNP 3 |
TGCAGATGCATTAAGGATAAATAATTATATTGTTTAGGCCT |
61 |
G>A |
0.989247 |
Vietnam |
ACATTACGACATTAATATATGGAACCCT |
|||||
SNP 4 |
TGCAGCTCTGGAAACACAGCACTATACAGCACGTCCTAAT |
31 |
C>T |
0.989247 |
International |
AATAGTAAAAGCAAGCCATGTAATGGAAA |
|||||
SNP 5 |
TGCAGGCTCCACTGCTGTTCATGTTGGCGAGGAGATTCAC |
45 |
G>A |
0.989247 |
Vietnam |
TCTGTACGTGTCTGGAGGATGATCTGCTG |
|||||
SNP 6 |
TGCAGAGTGACATTAGGTTATGTTGTATTTGCTTATCATAG |
62 |
C>G |
0.989247 |
International |
ACCTGTATCAGGGGTTCCCCT |
|||||
SNP 7 |
TGCAGTACTACAGCTACAATCCACTCAGCCACAGCACCAC |
50 |
C>A |
0.989247 |
International |
TGGGCAGCAACAGTGTGATTGCTGTGCTG |
|||||
SNP 8 |
TGCAGCTCTCTGTGTCCATGTTTCTCATTTTTTTGTGAACTA |
50 |
C>T |
0.978495 |
Vietnam |
AAGAGATTCGGCTGACTTGTGTATTCC |
|||||
SNP 9 |
TGCAGCTCTCGCTCTCAAATTCAAATGAGCCTTACTGGCAT |
66 |
T>G |
0.978495 |
Vietnam |
GGCGCTAGCAAGATACAGCATTTGGGAA |
|||||
SNP 10 |
TGCAGCAATTTTCCTCATAACATCAGTGAGTACTGCTCCTG |
58 |
C>T |
0.978495 |
International |
GAGTACACATAATGTTTCAAAAATGTGC |
|||||
SNP 11 |
TGCAGGTACTTAGGCTATTTGGGCTCCACATTCAGTTAAAA |
14 |
C>T |
0.967742 |
International |
TTCAGTTAAAGTTATTAAAAT |
|||||
SNP 12 |
TGCAGTCATCCACGGCTTCTGGTTGGAGCGTGATGTGATGG |
56 |
C>T |
0.967742 |
Vietnam |
TCTTGGAGGCGGTGACGTCATCAATGCA |
|||||
Table 2 The sequencing of SNPs location in Vietnamese and international catfishes.
Analysis of significant SNPs
Statistical analysis was performed using GraphPad Prism5 (GraphPad Software Inc., La Jolla, CA). SNPs with significantly different frequency ratios between Vietnamese catfish and international catfishes population were analyzed by one-way ANOVA. A p value of <0.05 was considered statistically significant.
Identification SNPs among populations
A total of potential 16,689 SNPs were observed at the lowest criteria setting. At the next selected set of criteria, minimum read depth is set as excluding the top 5% of minimum reads for SNP detection and maximum read depth is set as excluding top 5% of maximum reads for SNP detection, a total of 11,009 putative SNPs were identified, in which 780 SNPs observed in Vietnamese catfishes and others in international catfishes. Finally, we compared all putative SNPs between two strains and received twelve specific SNP markers (Table 2).
SNPs validation in Vietnamese catfish populations
Twelve special SNP markers selected from the preliminary Vietnamese catfish genome assembly map that confirmed by both PCR method using primers designed as (Table 1) and Sanger sequencing. Special SNPs identified from each of the area were shown in (Figure 1) (Figure 2). As shown in Figure 1, the primers designed for the first PCR successfully produced the desired products, except primers for SNP 6, 7, and 8 that failed in primer design and data not showed here. Analysis of the results showed that SNP 1, 2, 3, 4, 10, 11, and 12 appeared in the test regions, were different with the expected analysis. As shown in (Table 2), some SNPs (e.g., SNP 1, 2, and 3) only appeared in Vietnamese catfish samples without the presence of international catfish samples, however, the Sanger sequences showed that they have appeared in the samples of both areas. Similarly, SNP 4 and 11 should only appear in international catfish samples and not appear in Vietnamese catfish, but the results have been shown the opposite data. Other SNPs (e.g., SNP 10 and 12) did not show the mutation position like analysis results in (Table 2). Only two SNPs 5 and 9 were observed a similar data, these mutants appear only in Vietnamese catfish, but not in international samples. Thus, by re-checking the position of the mutant via PCR method and sequencing analysis, the SNPs 1, 2, 3, 4, 10, 11, and 12 were not used to determine the difference between catfish’s Vietnam and international catfish (Thailand, Cambodia, and Bangladesh). In this case, the observation of both SNP 5 and SNP 9 have been corrected from both PCR and sequencing data, and may be considered as special SNPs for Vietnamese catfishes.
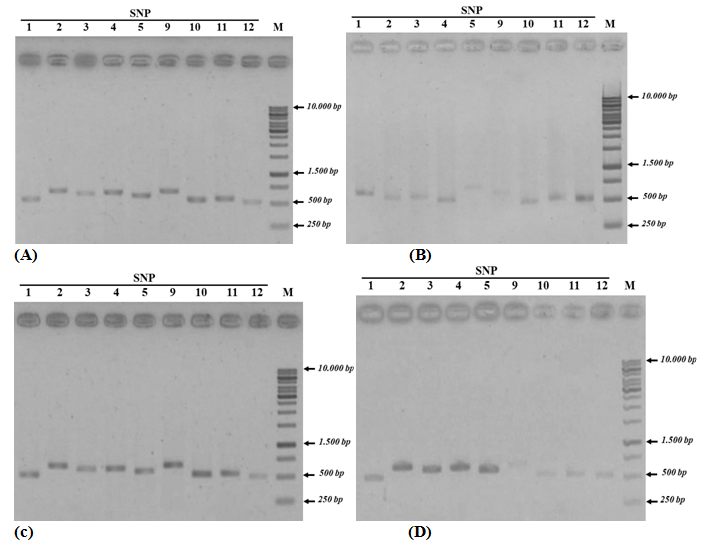

The number of each specific SNPs (e.g., SNP 5 and 9) was confirmed with gel electrophoresis in 300 individual Vietnamese catfishes (including 14 locations of 8 provinces) and 100 other individual international catfishes (e.g., 10 locations in 3 countries as Cambodia, Thailand and Bangladesh). The results were shown in (Table 3). Interestingly, 100% SNP5 observed in Vietnam catfish samples, whereas no expression in international catfish samples. Another SNP 9, approximately 75% - 84.4% of this SNP was identified in wild and farming catfishes, respectively. A significant difference (P <0.01) was appeared when validating the occurrence of SNP5 and 9 in both catfish populations, its mean may be expected that SNP 5 and 9 are specific for Vietnamese catfishes.
Location |
SNP 5 |
SNP 9 |
||
|---|---|---|---|---|
Wild catfish |
Farming catfish |
Wild catfish |
Farming catfish |
|
Vietnam |
300/300 (100%) |
300/300 (100%) |
75/100 (75%) |
76/90 (84.4%) |
International (Cambodia, Thailand, Bangladesh) |
0/30 (0%) |
0/60 (0%) |
4/30 (13.3%) |
6/60 (10%) |
P value |
0.0001(P< 0.01) |
0.0001(P< 0.01) |
0.0001(P< 0.01) |
0.0001(P< 0.01) |
Table 3 The occurrence of SNP 5 and SNP 9 in wild and farming catfish samples between Vietnam and the international regions
In recent years, several studies have successfully used NGS technologies for SNP marker discovery in fish species relevant to fishery and aquaculture, for instance, catfish,15 lake sturgeon,16 rainbow trout,17,18 lake whitefish,19 Atlantic cod,20 salmonids,17,18 hake,21 turbot,22 Atlantic herring23 and Pacific herring.24 According to the latest statistics on fish species, approximately 7,800 species (25%) have successfully researched at least one characteristic barcode for the species and the original data related to the species was deposited in BOLD.25 In the study, we have identified 12 specific SNPs (about 0.1 %) in the Vietnam catfish and the international catfish samples. The small numbers of SNPs detection may have been a result of sequencing the small population sizes. Besides, the catfish of four countries have not yet published their reference genome sequencing, therefore, when assembling and discovering SNPs, we used all their assumption genome sequencing of four catfish species. This might lead to the assembly of the same contig due to their high sequence similarity, single nucleotide differences between duplicated loci in the genome but invariant at the population or species level26 or single nucleotide variants with complex characteristics due to polymorphisms within duplicated regions,26 and all of situations were indistinguishable from SNPs during filtration. Additionally, the SNPs identified in this study using stringent criteria in the construction of special SNPs.27 Like Chao Li et al.28 studied in 190 individual fishes of five wild and domesticated populations (e.g., Mississippi River, Missouri River, D & B, Rio Grande and Texas) scored for 4,275 SNPs across all five populations. The confirmation these SNP in five populations only showed a total of 64 SNPs (1.5 %) that have successfully emerged in 100% individual in all populations.28 Another study found about 6.6 million, 5.3 million, 4.9 million, 7.1 million and 6.7 million common SNPs in Marion, Thompson, USDA103, Hatchery and wild fish populations, respectively, using the new generation sequencing system. Approximately 6% of all SNPs are identified in individual species.9
SNP combined with sequencing is also known as a useful indicator in the traceability of some aquatic species. Identification of specific SNPs of Vietnamese catfishes to avoid trade frauds in aquaculture and export between Vietnamese catfishes and Pangasius in the surrounding areas. We identified two SNPs appearing at levels of 75-100% in the catfishes of Vietnam, whereas only 0-13.3% in the international catfish samples. The other studies have identified SNPs in population that showed the similar parameters. Iratxe Montes et al.10 used the NGS sequencing technique to detect and confirm SNPs in European anchovy species without reference sequence. From the analysis results for the assumed 2,317 SNPs suitable for the design of priming tests in populations. A set of 530 individuals were tested, resulting in 83.2% of individuals presenting these SNPs.10 Neilsen et al.29 using genetic polymorphisms have found the origin of four commercial marine fish populations (e.g., Snowy Fish, Trichoderma fish, Trilobites and Coalfish) on European scale. The results were reported that 93-100% of individuals able to accurately retrieve origin via SNPs combined with Bayesian statistical methods.29 DNA barcodes are also used to detect fraudulent foods, including fish products.30 Recently, many cases of tuna identification have found misprinting and illegal trade.31 Yoon et al.32 developed a DNA chip to identify nine species of salmonids and managed the origin and commercial traceability of salmon.32 For acacia fishes, traceability of products by high molecular indicators avoids commercial fraud.33 From our results suggested that SNP5 and 9 can be used to make the bar-coding sequence distinguish between Vietnamese and international catfishes in the future.
Catfish species are almost impossible to distinguish based on phenotypes, therefore, the number of SNPs identified from this study can be potentially used for species identification, tracing the origin of commercial species, analyzing the genetic difference, and marking fish for other genetic experiments in Vietnamese catfish. The further, it will be useful for the development of high density SNP arrays for genetic and genomic analysis in catfish.
This research project was sponsored by Vietnam Ministry of Agriculture and Rural Development.
The author declares no conflict of interest.

©2018 Bich, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.