MOJ
eISSN: 2574-819X


Research Article Volume 2 Issue 1
1Department of Animal Science, Bharathidasan University, India
2Department of Biochemistry, Madurai Kamaraj University, India
3Department of Chemistry, Korea University, South Korea
4Lead Discovery Group, Discovery Biologics, Chigene Biotech Private Limited, India
Correspondence: Prakash Shyam Karuppiah, Lead Discovery Group, Discovery Biologics, Chigene Biotech Private Limited, Sivagangai TN 630211, India
Received: January 24, 2018 | Published: February 12, 2018
Citation: Venkatesan R, Prabhakaran R, Madar IH, et al. Theoretical designing of acetylcholine esterase (Ache) inhibitors. MOJ Biorg Org Chem. 2018;2(1):27-30. DOI: 10.15406/mojboc.2018.02.00051
In this manuscript, we have presented the results of binding efficacy of a range of chemical compounds that were generated from a modeled pharmacophore based upon structure clustering using donezaphil as a lead molecule. On analyzing the structure cluster result, we found that 2-((4-benzylcyclohexyl) methyl)-2,3-dihydro-1H-indene-5,6-diol is present common in all the active drug/potential lead molecules. Furthermore, we have established a structure activity relationship and studied the efficacy of the ligands generated. Our results indicate that the compounds generated based on the pharmacophore model preferentially binds to the allosteric site of AChE. The binding energy calculation of all 28 compounds that were generated from the modeled pharmacophore reveals favorable interaction with moderate to low binding energy relative to donezaphil. Among the 28 compounds generated, F54 was found to be the most effective compound based upon in silico binding prediction, total energy and toxicity considerations. Hence we propose to synthesis and further evaluate F54 that may show interesting biological activity. In addition, we have validated a hypothesis with evidence that the ligand targeted for AChE must possess a polar side group, preferentially NH3+. The pharmacphore generated and presented in this study may serve a backbone structure for future ligands targeting AChE.
Keywords: alzheimer’s disease, acetyl cholinesterase, pharmacophore model, toxicity analysis, drug compound, pathophysiological condition, donepezil, cerebral cortex, nucleus basalis, hippocampus, ligand atoms, docking calculations, tanimoto, dielectric functions, non-polar hydrogen atoms
SAR, structure activity relationship; AChE, acetyl cholinesterase; SLA, single linkage algorithm; ST, similar shape
Alzheimer’s disease still remains as an untreatable neurological disorder with no successful treatment strategy to prevent or cure or reverse the damage. Globally, AD is 5th major cause of death. Over last three decades, accumulating evidence for Alzheimer’s suggest multiple causes, predominantly the gene make up of affected individuals are altered. Affected individuals suffer from severe loss in brain weight and volume. AD is also characterized by increase in neuronal populations in some regions of brain than the other areas. The loss of neuron is sought to be one of the reason to cause loss of brain volume. However, previous studies have reported a strong correlation of acetylcholine deficiency and memory loss along with cognitive deficits of patients suffering from AD. Acetyl cholinesterase (AChE) is reported to be a key enzyme in AD. Over decades in Alzheimer's drug research, inhibitors of AChE remain to be of prime focus. Despite substantial efforts in designing efficient drug that can either treat or halt the progression of AD, none of them appears to be fully effective until now. Of note, the drug market for managing AD in 2010 alone is 604 billion dollars.1 The pharmaceutical industry continues to seek new molecule that could improve the treatment strategy for AD at lower cost. Herein, we attempt to identify potential lead molecule for AD treatment by virtual drug screening approach. We employ structure activity relationship (SAR) to develop a pharmacophore model;2 from the basic phamacophore structure we have developed a database with potential lead molecules that could inhibit AChE.3–5 This report discusses the binding behavior of lead molecule from the generated chemical database using suitable computational methods. In addition, we also report the experiment results of toxicity analysis, prediction of active spectra for biological activity that includes pharmacological effects, predicted mechanisms of action, interaction with metabolic enzymes influence on gene expression. Virtual design and analysis allows us to understand and perform preliminary screening for a complete set of novel compounds whose experimental data is not available in literature till date.
Structure clustering
In order to define a common backbone structure for further modeling of the pharmacophore we performed tanimoto similarity search of available drug compound with Donepezil as model drug compound. Donepezil is one of the most widely used drugs for AD that reversibly inhibits AChE.6 It is found to increase cortical acetylcholine on therapeutic use.7 As we discussed early in the introduction acetylcholine is correlated with the pathophysiological condition of AD especially the loss of memory function and cognitive deficits is improved by increase in acetylcholine. Therefore inhibiting AChE would effectively increase the acetylcholine level in cerebral cortex, nucleus basalis, and hippocampus. Structurally, Donepezil is a piperidine derivative and differs greatly from other AChE inhibitors.8 Also, clinical evidence suggest that Donepezil selectively inhibits AChE in brain that provides a strong rationalé to search for drug compound that is related to donepezil, still effective than donepezil.
The compounds that are structurally similar to Donepezil are clustered using the single linkage algorithm (SLA algorithm for substructure search).9 The structure similarity is reported as tanimoto score that is calculated from the 2D structure fingerprint. Theoretically, a tanimoto score of 0.68 or greater is statistically significant at the 95% confidence interval. In addition to tanimoto similarity search we have also performed 3D structure similarity search, a similarity score is of 0.88 is considered as statistically significant.10 The algorithm searches all the known drug compounds for 2D structure analogs, and 3D structures from PubChem database (Figure 1). Figure 2A shows the tanimoto similarity structure cluster for selected conformers that satisfy the tanimoto score, the conformers were paired by most similar structures with 3D superposition optimized by most similar shape (ST).11 The 3D similarity is aligned by shape (ST) and feature (CT) with ten conformers per compound (nconf). Tanimoto similarity search results in 42 structurally similar compounds that could exert similar activity pharmacologically. On analyzing these 42 compounds we found 2 common backbone structures (Figure 2B and 2C). In our opinion, this backbone structure could be one of the factors that aid the binding of AChE protein in brain. The results of tanimoto similarity search are tabulated separately in supplementary Table 1.
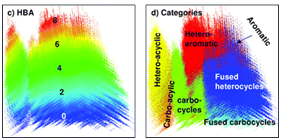
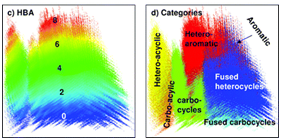
Figure 1 Descriptor analysis of the modeled pharmacophore using tanimoto similarity search algorithm for
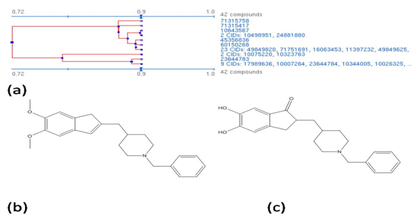
Figure 2
In order to define a pharmacophore, a deeper understanding of the structure is critical, on analyzing the above two motifs we found that 2-((4-benzylcyclohexyl) methyl)-2,3-dihydro-1H-indene-5,6-diol (C23H28O2) is present common. Hence, we use the structure as backbone for our pharmacophore model.
Pharmacophore modeling and AChE docking
After generating a pharmacophore model we generated a range of potential active lead compounds by modifying their side chains. The compounds that were subjected for analysis were listed in Figure 3. Docking calculations were carried out using Swiss Server.12 Gasteiger partial charges were added to the ligand atoms. We merged the non-polar hydrogen atoms and defined the rotatable bonds prior to simulation. After preparing the protein and ligand, we simulated the binding energy calculation for AChE protein and pharmacophore model. The essential hydrogen bonds, charges and solvation parameters were added to the AChE model. We first developed an affinity (grid) maps of 10Å grid points with 0.375Å spacing.13 In calculating the van der Waals and the electrostatic terms we used parameter set- and distance-dependent dielectric functions, respectively. Visualization and analyses were performed with various tools that including Chimera 1.8.1, Discovery studio visualizer, and PyMOL.
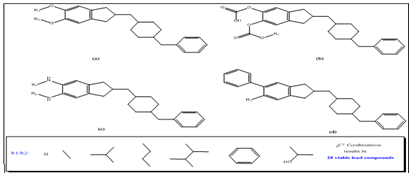
Figure 3 Backbone structures used to generate the chemical library; the functional groups R1 and R2 are selected in combinatorial fashion and are shown in as figure inset.
The docking algorithm is developed in such a way that multiple binding modes are generated either in a box to perform local docking or in the vicinity of all target cavities to perform blind docking. Simultaneously, we estimate their CHARMM energies on a grid.14 We further clustered and evaluated the binding modes with the most favorable energies. Lamarickian genetic algorithm was used to perform docking. In addition, we encoded a command to release the rotatable torsions while docking. However the initial position and orientation of the protein was set randomly. After computing 250000 energy values with population size about 150, a translational step of 0.2Å was applied.15
The ligands were selected based upon the combinatorial method. Figure 3 shows the backbone structure and functional group that varies among the ligands. A total of 28 viable lead compounds were identified and binding mode was predicted for these compounds. The results of binding mode and subsequent analysis were shown in Figure 4. A potential lead compound has the lowest binding energy, among the compounds investigated compound F54 showed relatively lower binding energy and better interaction with AChE. As shown in Figure 5 there are few compounds that showed favorable interaction with AChE however none of them bind directly to active site residues. Most of the binding occur at distant sites other than the active site, although there is possible allosteric mode of inhibition, the efficacy of a drug with such mechanism is still questionable. Since we do not have any experimental data for these molecules nor the molecules have been synthesized, it would be wiser if we could pick up a lead compound with competitive mode of interaction that fits directly into the active site pocket resulting in the inhibition of substrate binding.
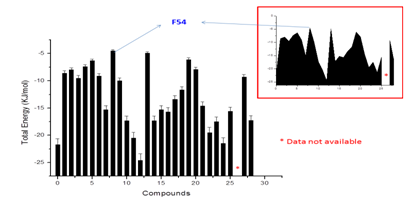
Figure 4 Bar plot showing the results of calculated total energy (ΣE) in KJ/mol for individual compounds. Values are expressed as ΣE±SEM.
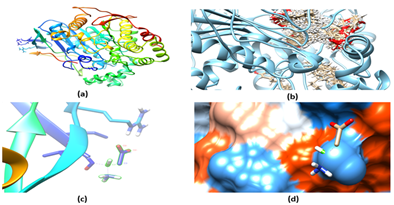
Figure 5
The results of our docking calculation indicated that N-terminal amino acids majorly participate in the interaction of potential ligand (F54) (Figure 5D), this may be owing to the fact that the amino side chain of the ligand forms hydrogen bonding with the polar amino acid resulting in more stable complex than the other ligands that were investigated herein, thus it could be generally stated that introducing polar compounds increases the binding efficacy. On the other hand, increasing the aromaticity results in poor binding energy (Figure 4). Therefore it is suggested to include a polar side groups to improve the binding efficiency while synthesizing inhibitors for AChE. From the binding energy calculation and theoretical analysis it can be interpreted that the modeled pharmacophore can be used effectively to synthesize potential inhibitors for AChE for effective management of Alzheimer’s disease.
Tanimoto similarity search and structure clustering
Tanimoto similarity search was performed of available drug compound from PubChem database. Donepezil was used as model drug compound. The compounds that are structurally similar to Donepezil are clustered using the single linkage algorithm (SLA algorithm for substructure search).9 The structure similarity is reported as tanimoto score that is calculated from the 2D structure fingerprint (tanimoto score≥0.68, P>95%). 3D structure similarity search was performed for selected compounds (Similarity Index>0.88, P>95%).10 We defined the pharmacophore by identifying common structural motif and generated a range of potential active lead compounds by modifying their side chains.
Docking studies
Docking calculations were carried out using Swiss Server.12 Prior to docking calculation, Gasteiger partial charges were added to the ligand atoms. The non-polar hydrogen atoms and the rotatable bonds were defined in ligands prior to simulation. Similarly, the essential hydrogen bonds, charges and solvation parameters were added to the AChE model. Affinity (grid) maps of 10Å grid points with 0.375Å spacing was developed.13 Parameter set- and distance-dependent dielectric functions were used for vander Waals and the electrostatic terms, respectively. Visualization and analyses of the protein were performed using Chimera 1.8.1, Discovery studio visualizer, and PyMOL.
Lamarickian genetic algorithm was used to perform docking. Multiple binding models were generated either in a box to perform local docking or in the vicinity of all target cavities to perform blind docking. CHARMM energies on a grid were determined.14 The rotatable torsions were released while performing docking. The initial position and orientation of the protein was set randomly. After computing 250000 energy values with population size about 150, a translational step of 0.2Å was applied.15 The ligands were selected based upon the combinatorial method.
Treatment of Alzheimer’s disease remains challenging task. Based upon our theoretical calculations we found that 2-((4-benzylcyclohexyl) methyl)-2,3-dihydro-1H-indene-5,6-diol could serve as an phamacophore model to synthesize potential lead molecules. In addition, based upon binding energy calculation we hypothesize that the lead molecules generated via this phamacophore model may preferentially bind to the allosteric site of AChE and the ligand targeted for AChE must possess a polar side group, preferentially with amine (NH3+) functionality. We had generated 28 compounds based upon this phamacophore, among them F54 was found to be the most effective compound based upon in silico binding prediction, total energy and toxicity considerations. In conclusion, the pharmacophore generated and presented in this may serve a backbone structure for future ligands targeting AChE therefore we propose to synthesis and further evaluate F54.
The authors greatly acknowledge Mr. K. Prakash Shyam for his valuable insights into this manuscript. There is no potential conflict of interest in terms of funding.
The author declares no conflict of interest.

©2018 Venkatesan, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.