MOJ
eISSN: 2574-9722


Research Article Volume 3 Issue 5
Department of Biotechnology, Stella Maris College, India
Correspondence: Aruna Sharmili S, Assistant Professor, Department of Biotechnology, Stella Maris College (Autonomous), Chennai, India
Received: July 05, 2020 | Published: November 30, 2018
Citation: Maria ABF, Sharmili AS, Anbumalarmathi J. Isolation and characterization of actinomycetes from marine soil. MOJ Biol Med. 2018;3(6):221–225. DOI: 10.15406/mojbm.2018.03.00103
Four different actinomycetes were obtained from the sampling sites. The isolate AA1 isolated from Puducherry beach exhibited good antibacterial activity on primary screening when compared to the others and hence was chosen for further study. The isolate AA1 was Gram positive with smooth surface morphology and the spore chain morphology showed them to be simple rectus. When the isolate was grown on different media the ariel mycelia was mostly white while the substrate was cream/white. It was able to grow at a pH of 7 at 30°C and tolerate 7% NaCl. Maltose, lactose and xylose was used as the sole carbon source. The crude ethyl acetate extract of AA1 showed antibacterial activity against E.coli (9mm), S.aureus (7.5mm), P.mirabilis (7.7mm), P.aeruginosa (7mm) and K.pneumoniae (4.5mm) at a concentration of 3.5mg/ml. MIC of the crude extract of AA1 was 0.531mg/ml for E.coli and P.aeruginosa, 1.062mg/ml for S.aureus, 0.265mg/ml for P.mirabilis, and 2.125mg/ml for K.pneumoniae. The FT-IR Spectrum of the crude extract showed characteristic peaks at 3290.56cm-1, 1635.64cm-1 and 1016.49cm-1 corresponding to the following functional group such as alcohol (O-H), Alkene (C=C), Ether (C-O) respectively. The 16S rRNA gene sequences of the isolate AA1 revealed 100% similarity to Streptomyces lincolnensis. The restriction sites prediction of the 16SrRNA gene of Streptomyces lincolnensis showed restriction sites for Eco53KI, Bsr I, Hae II, Eco RV, EcoRI, EagI, Hinc II etc. The restriction site analysis showed GC content of 59% and AT content of 41%.
Keywords: streptomyces lincolnensis, antibacterial activity, minimum inhibitory concentration, FT-IR
Streptomyces are the largest genus of Actinobacteria proposed by Waksman and Henrici in 1943 are Gram positive, aerobic, spore forming filamentous bacteria.1 They produce an extensive branching substrate mycelium that are tough, leathery and produce diffusible pigments.2 Their aerial mycelium bears chains of arthrospores. Streptomyces have genomes with high Guanine and Cytosine content (69-78%).3 They are the largest antibiotic producing genus and about 70% of the antibiotics have been isolated from them.4 They also produce other type of compounds that are deleterious to competing microbes5 apart from a wide array of natural bioactive compounds such as antifungal, antiviral, anti- hypersensitivity, antitumor.6,7 Bioactive compounds produced from Streptomyces are not only used in the treatment of human and animal diseases but also used as bio control agents.8 They are also known producers of industrial enzymes, agrochemical and pharmaceutical products.9 They play important role in carbon recycling, degradation of organic matter and also enhance soil fertility.10 Development of antibiotic resistance microbes to the commonly available antibiotics and antifungal agents has necessitated the requirement of new compounds and the members of the genus Streptomyces offers promising lead compounds.11 The present investigation was undertaken to isolate, screen and characterize Streptomyces from marine soil sample.
Collection of soil samples
Soil samples were collected from Stella Maris college campus and Puducherry beach. The samples were mixed along with calcium carbonate to reduce Gram negative bacteria and air dried for 10days. One gram of soil sample was added to 100ml sterile and left in the shaker for 12hours. Serial dilution was done up to 10-5 and plated on Actinomycetes Isolation Agar supplemented with 50µg/mL Streptomycin sulphate and 75µg/mL of Nalidixic acid. In the case of the marine sample plates were prepared with 50% sea water. The plates were incubated at 28°C for 14 days.12
Screening of antibacterial activity
The primary screening of pure isolate was determined by cross streak method in Nutrient agar plates.13 The isolates were streaked as a parallel line on Nutrient agar plates and incubated at 28°C for 5 days. After observing a good ribbon like growth, Escherichia coli (ATCC 25922), Staphylococcus aureus (ATCC 25923), Klebsiella pneumoniae (ATCC 700603), Proteus mirabilis (ATCC 25933) and Pseudomonas aeruginosa (clinical isolate) were streaked at right angles to the original streak of actinomycetes and incubated at 28°C for 24hours. In the secondary screening the mature spores of the isolate was inoculated in 100ml of ISP 2 broth and incubated at 30°C for 15days.13 The culture broth was centrifuged at 5000rpmat 4°C for 20min and the supernatant was collected. 100µl (10-5cells) of E. coli, S. aureus, K. pneumoniae, P. mirabilis, P. aeruginosa were plated on Muller Hinton agar plates. Wells were made using sterile cork borer and the culture supernatant was added. Streptomycin sulphate (1µg) was taken as the positive control.
Morphological, physiological and biochemical characteristics
Cultural characteristics of the isolate was studied based of the intensity of the growth, growth pattern, colony colour along with colour of aerial mycelia on Trytpone yeast agar (ISP medium 1), Yeast extract malt extract agar (ISP Medium 2), Oat meal agar (ISP Medium 3), Inorganic salt starch agar (ISP Medium 4), Glycerol Asparginine agar (ISP Medium 5), Peptone yeast extract iron agar (ISP Medium 6), Tyrosine agar (ISP Medium 7) as described by Shirling and Gottlieb, 1966 and also on Starch casein agar, Potato Dextrose agar, Kuster’s agar and CzapekDox agar. Gram staining and spore surface morphology was examined by scanning electron microscopy (SEM). The arrangement of spore and sporulating structures were examined microscopically by using cover slip culture method.14
Physiological characteristics
The ability of the isolates in utilizing various carbon compounds was tested on ISP basal meium915 supplemented with a final concentration of 1% carbon sources such as maltose, fructose, xylose, sucrose and lactose. Biochemical tests such as Indole production, Methyl red (MR), Voges Proskauer (VP), Citrate utilization, starch hydrolysis, phenylalanine Deamination, Carbohydrate fermentation (arabinose, xylose, adonitol, rhamnose, cellobiose, mellibiose, saccharose, raffinose, trehalose, glucose and lactose), Triple Sugar-Iron agar test, nitrate and nitrite reduction were carried and the results were compared with Bergey’s manual of determinative bacteriology. Growth of the isolate in different concentration of sodium chloride (2.5%, 5% and 7%), different temperatures (20°C, 30°C, 45°C and 55°C) and pH (5.8,7 and 9) were also studied.13
Production of secondary metabolites by submerged state fermentation
250ml of ISP 2broth was inoculated with 2ml suspension of pure actinomycete isolate and kept for 15 days in a rotary shaker at 30°C.14 The fermented broth was centrifuged at 5000rpm at 4°C for 20min. the supernatant was collected and equal volume of ethyl acetate was added and shaken vigorously at regular intervals. The solvent phase was separated from the aqueous phase by using a separating funnel and evaporated at room temperature to remove the solvent. The crude extract thus obtained was stored.
Antibacterial analysis of the crude extract
The crude extract residue obtained was weighed and re-suspended in DMSO and a stock solution of known concentration was prepared and used for antimicrobial analysis.9 The antibacterial activities of the crude extract obtained from the submerged state fermentation was tested by using agar well diffusion method. 100µl (10-5 cells) of E. coli, S. aureus, K. pneumoniae, P. mirabilis, P. aeruginosa were plated on Muller Hinton agar plates. Wells were made using sterile cork borer and the culture supernatant was added. Streptomycin sulphate (1µg) was taken as the positive control.
Determination of minimum inhibitory concentration
1ml of Nutrient broth was dispensed into 10 test tube and 2ml into the 11th test tube (broth control). 2ml of the crude extract alone was added in the 12th test tube (crude extract broth). 1ml of crude extract solution was added to the test tube 1 and 2 and serial dilution was done from the 2nd up to the 10th test tube. Finally 1ml was discarded from the 10th test tube. 1ml inoculum (10-5 cells) of E. coli, S. aureus, K. pneumoniae, P. mirabilis, P. aeruginosa was added into the test tubes and incubated 37°C for 18-24hrs. After overnight incubation, the lowest concentration inhibiting the visible growth of the microorganism was considered as the MIC value.13
Characterization of crude extract by Infrared spectroscopy (FT-IR)
The infrared (IR) spectra of the crude ethyl acetate extract was measure (as KBr discs) between 300-4000cm-1 on Perkin Elmer 2000FT-IR spectrophotometer.13 The functional group and types of vibration were assessed based on the peak value.
Molecular characterization
The isolate AA1 was grown in ISP 2 broth and the total genomic DNA was extracted according to the protocol of Sambrook et al.16 PCR amplification were carried out in a final volume of 30µl o reaction mixture containing 3μl of template, 1μl of 2.5mM each dNTP’s master mix, 0.2μl of 5U/Taq DNA polymerase, 1μl each of 5 p.mol 16S rRNA Forward primer 27F and Reverse primer 1492R, 3μl of 10x Buffer and 20.8μl o sterile Millipore water. Amplification was carried for 30 cycles (95°C for 5min, 95°C for 30sec, 55°C for 30sec and 72°C for 2min and final extension at 72°C for 10min). Following amplification, 2μl of the PCR amplified products was run on a 2% agarose gel in 1X TAE buffer and the molecular sizes of amplicons were determined using a standard 1-kb DNA ladder. 30µl of the amplicon was purified by EZ-10 Spin Column, (BIO BASIC INC). The purified amplicon was sequenced by Big dye terminator cycle sequencing kit. The 16S rRNA gene sequence was aligned with the corresponding 16S rRNA sequence of the type strain of members of the genus Streptomyces retrieved from the GenBank using BLAST program (http://www.ncbi.nlm.nih.gov/nuccore/. The software package MEGA X was used for multiple sequence alignment17 and phylogenetic analysis.18 The phylogenetic tree was constructed using the maximum likelihood method. Individual branches were determined by bootstrap analysis based on 1000 sampling.
Restriction site analysis
The restriction sites of the DNA of the strain were analyzed by NEB cutter version 2.0 (http://tools.neb.com/NEBcutter2).
Isolation and screening of antimicrobial activity
Four different colonies of actinomycetes were obtained in the study. Two white powder colonies, one grey colony and a cream colony were obtained. Results of primary and secondary screening indicated that isolate AA1 (Aruna Alice) that was isolated from Puducherry beach soil exhibited better activity compared to the other three colonies and hence was taken up for further study (Figure 1). The isolate AA1 exhibited antimicrobial activity against E. coli, S. aureus, P. mirabilis, and P. aeruginosa but had no antibacterial activity against K. pneumoniae. In the secondary screening the isolate exhibited activity against K. pneumonia.
Morphological, physiological and biochemical characteristics
The isolate was Gram positive and the spore chain morphology was simple rectus with smooth surface as seen from SEM analysis (Figure 2).
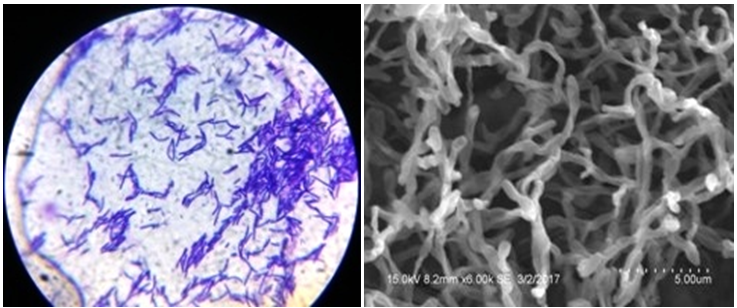
Figure 2 Gram staining (a) and Scanning Electron Microscope (b) morphology of actinomycete isolate AA1.
The aerial mycelium of actinomycete isolate AA1 was white in Nutrient agar, Starch casein agar, Kuster’s agar, Czapek Dox agar, ISP–1 to 4, ISP–6 where as it was cream in Potato Dextrose and Tyrosine agar (ISP-7). Grey white mycelium was observed on ISP–5. The substrate mycelium was cream colored on Nutrient agar, Czapek Dox agar, ISP-1 to 3. It was white in the case of Starch casein agar, Kuster’s agar, Potato Dextrose, ISP % to 7. It was pale white on ISP–4 (Inorganic salt starch agar) (Table 1).
Medium |
AA1 |
|
Aerial Mycelium |
Substrate Mycelium |
|
Nutrient agar |
White |
Cream |
Starch casein agar |
White |
White |
Potato dextrose agar |
Cream |
White |
Kuster’s agar |
White |
White |
Czapek dox agar |
White |
Cream |
ISP–1 Tryptone yeast extract agar |
White |
Cream |
ISP–2 Yeast malt extract agar |
White |
Cream |
ISP–3 Oat meal agar |
White |
Cream |
ISP–4 Inorganic salt starch agar |
White |
Pale white |
ISP–5 Glycerol asparagine agar |
Grey white |
White |
ISP–6 Peptone yeast extract iron agar |
White |
White |
ISP–7 Tyrosine agar |
Cream |
White |
Table 1 Cultural characteristics of Actinomycetes isolate AA1
The actinomycetes isolate AA1 able to grow on NaCl concentration of 2.5%, 5% and 7% but growth was more in 7% (Figure 3). The optimum pH for growth was 7. The organism did not survive the acidic pH of 5.8 and alkaline pH of 9 (Figure 4). The optimum temperature was 30°C though growth was observed even in 45°C but there was no growth in 50°C (Figure 5). Isolate AA1 was positive for urease, nitrate and nitrite reduction, Indole, methyl red, citrate and phenylalanine and negative for Voges Poskauer, TSI and starch. Maltose was utilized (++) followed by Lactose and Xylose (+). Fructose showed doubtful (±) utilization whereas sucrose as a carbon source was not utilized (-).
Antibacterial analysis and Minimum Inhibitory Concentration (MIC) of the rude extract
The crude ethyl acetate extract of AA1 showed antibacterial activity against E. coli (9mm), S. aureus (7.5mm), P. mirabilis (7.7mm), P. aeruginosa (7mm) and K. pneumoniae (4.5mm) at a concentration of 3.5mg/ml. In a previous investigation by Mathur et al.19 the ethyl acetate extract of Streptomyces sp was active against E.coli (20mm), P.aeruginosa (18mm), B. subtilis (17mm), P.vulgaris (13mm), and Klebsiella (5mm). MIC of the crude extract of AA1 was 0.531mg/ml for E. coli and P.aeruginosa, 1.062mg/ml for S. aureus, 0.265mg/ml for P. mirabilis and 2.125mg/ml for K. pneumonia (Table 2).
Test organisms |
Zone of inhibition (mm) |
MIC (mg/ml) |
|
Positive control (Streptomycin (20mg)) |
Crude extract (3.5mg) |
||
E.coli |
11 |
9 |
0.531 |
S.aureus |
10 |
7.5 |
1.062 |
P.mirabilis |
7.5 |
7.7 |
0.26 |
P.aeruginosa |
8.5 |
7 |
0.531 |
K.pneumoniae |
7 |
4.5 |
2.125 |
Table 2 Antibacterial activity and MIC of the crude extract of AA1
FT-IR analysis of the crude extract
The FT-IR spectrum of the crude extract of AA1 showed characteristic peaks at 3290.56cm-1, 1635.64cm-1 and 1016.49cm-1 corresponding to the following functional group such as alcohol (O-H), Alkene (C=C), Ether (C-O) respectively (Figure 6). Presence of O-H suggests the presence of phenolic compound in the extract. Phenolic type antimicrobial agents are known for their disinfectant, antiseptic properties.7
Molecular characterization
The 16S rRNA gene sequences of the isolate AA1 showed 100% identity to Streptomyces lincolnensis and was retived from GeneBank database using BLAST. The sequence was deposited in the GenBank and accession number of MG002523 was obtained. The phylogenetic tree was constructed using MEGA X with Nocardiopsis alba strain PC 2702 as the outgroup (Figure 7). Using the Maximum Likelihood method based on Tamura 3-parameter method the evolutionary history was inferred.20 The tree with the highest log likelihood (-5067.99) was considered. The percentage of tree in which the associated taxa clustered together is shown next to the branches. The tree was drawn to scale, with branch lengths measured in the number of substitutions per site. 41 nucleotide sequences were involved and all positions with less than 95% site coverage were eliminated. There were a total of 1348 positions in the final dataset. Evolutionary analyses were conducted in MEGA X.21
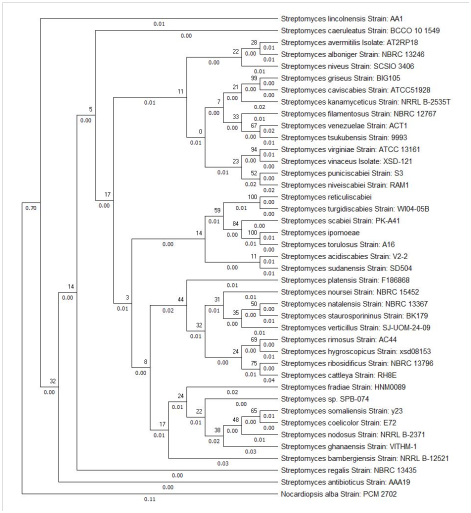
Figure 7 Molecular Phylogenetic analysis by Maximum Likelihood method using MEGA X showing the evolutionary relationship of Streptomyces species.
Restriction site analysis
The restriction site predictions of the 16S rRNA gene of Streptomyces lincolnensis showed restriction sites for Eco53KI, Bsr I, Hae II, Eco RV, EcoRI, EagI, Hinc II etc. the restriction site analysis showed a GC content of 59% and AT content of 41% (Figure 8).
The isolate AA1 was identified as Streptomyces lincolenesis. It exhibited maximum growth at pH 7 at 37°C and was able to tolerate 7% NaCl. Results of the antibacterial activity exhibited by the crude extract of the isolate indicates the presence of potent bioactive compound. Further study needs to be carried out to identify and investigate the efficacy of the bioactive compounds.
The crude extract of the isolate exhibited good antibacterial activity against the test bacteria used in the study. Results of the present investigation indicate that the actinomycete isolate AA1 can be used for the isolation of bioactive compound that can be used as a drug.
Authors thank the principal and the management of Stella Maris College (Autonomous), Chennai, Tamil Nadu, India, for the research facilities provided. The authors also thank Ms Vinetha Arul Mary B for helping us with the Bioinformatics tools.
The authors declare no conflicts of interest.

©2018 Maria, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.