MOJ
eISSN: 2573-2951


Mini Review Volume 4 Issue 1
1Department of Pharmacology and Chemical Biology, Baylor College of Medicine, USA
2Department of Pathology and Immunology, Baylor College of Medicine, USA
Correspondence: Damian W Young, Department of Pharmacology and Center for Drug Discovery, the Baylor College of Medicine, 1 Baylor Plaza, Houston, TX 77030, USA, Tel 713-798-5419
Received: September 20, 2017 | Published: November 3, 2017
Citation: Chang L, Young DW. Using fragment based drug discovery to target epigenetic regulators in cancer. MOJ Bioequiv Availab. 2017;4(1):223-230. DOI: 10.15406/mojbb.2017.04.00062
Cancer is associated with epigenetic changes such as abnormal DNA methylation and histone tail modifications, and these changes result in tumor suppressor gene silencing, oncogene overexpression, and genome instability – phenomena that are closely linked to cancer development. The epigenetic marks on DNA and histones are reversible and therefore it is possible to restore normal patterns through chemically targeting epigenetic regulators that generate, maintain, recognize, and remove these marks. In recent years, fragment–based drug discovery (FBDD) has been used to target both the protein–protein interactions (PPI) as well as the enzymatic functions of epigenetic factors. Low molecular weight fragments (MW<300) efficiently sample chemical space and can bind to discreet binding pockets on target proteins such as those found on PPI interface. In this review, we describe the biophysical, biochemical, and in silico methods used for FBDD. We also discuss the examples of using FBDD to target epigenetic readers such as bromodomain–containing proteins and epigenetic enzymes such as arginine methyltransferases 6 (PRMT6), lysine demethylase 4 (KDM4), and Sirtuin 2 (SIRT2). FBDD provides an economical alternative to traditional high throughput screening. Effective FBDD endeavors depend heavily on gaining structural information of ligand–protein interactions early on and the close collaboration between biophysicists, chemists, and biologists in all stages of the drug discovery process. Using FBDD, protein–protein interactions in non–enzymatic epigenetic factors can potentially be chemically modulated. Furthermore, FBDD enables the identification of new binding pockets that can improve compound specificity against various epigenetic enzymes.
Keywords: fragment–based drug discovery, epigenetic, cancer, screen, bromodomain, biophysics, histone
FBDD, fragment–based drug discovery; PPI, protein–protein interactions; HTS, high throughput screening; LE, ligand efficiency; SPR, surface plasmon resonance; NMR, nuclear magnetic resonance spectroscopy; SPR, surface plasmon resonance; TSA, thermal–shift assays; TR–FRET, time–resolved fluorescence energy transfer; ALTA, anchor–based library tailoring; BRD4, bromodomain containing 4; DSF, differential scanning fluorimetry; DNA, deoxyribonucleic acid; MD, molecular dynamic; SAM, s–adenosyl–l–methionine; SP, scintillation proximity
Cancer has traditionally been viewed as a genetic disease associated with the mutation or deletion of tumor suppressor genes and the mutation or amplification of oncogenes drives cancer development. However, over the past decade, mounting evidence has shown that epigenetic changes play crucial roles in tumor suppressor gene silencing, oncogene overexpression, and genome instability.1,2 Epigenetic modifications are heritable changes within DNA, protein, or RNA that are not associated with direct mutations within genomic DNA sequences.1 The first epigenetic abnormality in cancer to be recognized was the DNA methylation of cytosine to 5-methyl cytosine.3,4 In cancer genomes, highly repetitive regions become hypomethylated, while the CpG islands that are located upstream of genes become hypermethylated; completely in reverse of the patterns of normal cells.4 DNA hypomethylation in the repetitive regions loosens up chromatin structures, facilitates the binding of transcription factors, activates previously dormant transposons and retrovirus, and leads to genome instability.5 On the other hand, hypermethylation of CpG islands condenses chromatic structures and can inhibit tumor suppressor gene expression.3 Aside from DNA methylation, another prominent epigenetic change in cancer is the aberrant modification of histones.6 The genomic DNA is wrapped around histone proteins to form nucleosomes, and the acetylation, methylation, phosphorylation, ubiquitination, and sumoylation of various histone tails (the ‘histone code’) regulate the chromatin structure, transcription factor binding, and gene expression of any genomic locus.1,2,6 Cancer cells harbor abnormal histone modifications due to the mutation, silencing, or overexpression of various epigenetic regulator proteins that are responsible for the reading, writing, or erasing of histone marks associated with normal cell function.1,2 These alterations in the epigenetic marks on both DNA and proteins result in global changes of gene expression profiles that are linked to cancer development and progression.1,2,6 However, unlike genetic mutations, epigenetic modifications are generally reversible. Through the pharmacological targeting of various epigenetic factors, it is possible to re-establish normal epigenetic patterns on DNA and histone proteins, and thereby restore tumor suppressor gene expression and inhibit cancer cell proliferation.
Epigenetic marks on chromatin and DNA are generated, recognized, maintained, and removed by a plethora of epigenetic regulators. For instance, the DNA methyltransferases (DNMT1, DNMT3a, DNMT3b) catalyze the methylation of the 5-position of cytosine residues in DNA,7 while DNA demethylases (TET1, TET2, and TET3) catalyze the removal of methyl group.8 The histone tail modifications are much more diverse and therefore are catalyzed by a long list of enzymes. Many of these enzymes when deregulated are implicated in cancer development, including histone methyltransferases (ex. EZH2, SETD2, MLL, NSD1, NSD2, and NSD3),9 histone demethylases (ex. KDM1, KDM4, and KDM6A),9 histone acetyltransferases (ex. p300, CBP, and TIP60),10 histone deacetylases (ex. HDAC1, HDAC2, and HDAC8),11 and histone phosphatases (ex. Aurora Kinase B).12 Once established, the histone marks will bind to specific epigenetic readers that recruit other protein factors to translate histone codes into downstream effects.13,14 The epigenetic readers contain domains that recognize specific histone modifications: lysine methylation (ex. MBT, Tudor, PWWP, Chromosome, PHD, ADD, Ankyrin repeats, and WD40 domains), arginine methylation (ex. ADD, Tudor, and WD40 domains), lysine acetylation (bromodomain, DBD, DPF, double PH domains), and serine/threonine phosphorylation (14-3-3 and BIR domains).13,14 Many epigenetic regulators are overexpressed, silenced, or mutated in cancer cells and act as drivers for cancer development.2 Therefore, in recent years, there has been a significant amount of research both in academia and pharmaceutical companies aimed at modulating oncogenic epigenetic regulators to develop novel cancer therapeutics.2 This review will focus on the strategies of fragment-based drug discovery for generating small-molecule epigenetic modulators.
There are different challenges associated with the development of small-molecule epigenetic regulators given that they may be enzymatic or non-enzymatic. Epigenetic enzymes are more traditional drug targets and inhibitors can be developed through highly established processes such as high throughput screening (HTS),15 in silico screening,16 and rational structural-based drug design.17 In HTS, a collection of ~1 million compounds is screened against the protein target mostly by an enzymatic assay and less frequently by simple biophysical binding assays (ex. thermal shift, surface plasmon resonance).15 A well-resolved protein structure from X-ray crystal diffraction or NMR analysis is generally required for in silico screening.16 Using specialized software, large libraries of commercially-available compounds can be virtually docked one-by-one into the target structure’s active site.16 Subsequent to the computational screen, hits can be purchased or synthesized and then assayed for activity against the epigenetic enzymatic target. Finally, in rational structure-based design, the co-crystal structures of an enzyme and its substrate (ex. DNA and histone) or co-factors (ex. S-Adenosyl methionine and NAD+) are utilized as starting points for the design of competitive inhibitors.17 However, due to the conservative nature of enzymatic catalytic domains, the main challenges for targeting epigenetic enzymes are to ensure compound specificity among same enzyme family members.2
Many epigenetic factors, such as histone mark readers, lack enzymatic activities and operate mainly through protein-protein interactions (PPI) to recognize specific epigenetic marks and recruit other protein factors.2,13,14,18 These non-enzymatic epigenetic factors (ex. BRD4, ING1, and PHF6) contribute to abnormal gene expression programs and thus participate in cancer development making them interesting drug targets.2 However, targeting PPIs has proven to be more challenging than targeting enzymes leading to the prevailing notion that they are not “druggable”.19,20 The level of difficulty in developing PPI inhibitors is depends on the type of the two interacting proteins. For example, the interaction between two globular proteins is most difficult to perturb due to the large and flat protein interfaces that characterize their binding which lack large pockets for small molecules to bind.20 In contrast, the interaction between a globular protein with an anchor residue on peptide, such as a methylated or acetylated lysine on histone tails, are much more druggable, given that they contain deeper binding pockets responsible for recognizing the peptide modification.20 Toward the goal of developing small-molecule probes and lead-like compounds for each of these types of PPI epigenetic targets, advances in biophysical techniques and the application of fragment-based drug discovery (FBDD) have greatly improved success.20-22
FBDD is a complementary strategy to HTS and is a validated approach toward generating small-molecule lead-like compounds and FDA approved therapeutics. The central theme underlying FBDD is the screening of a library of low molecular weight compounds (<300g/mol), or “fragments,” against a specific biological target.22 The small size of fragments enhances the hit rate against target proteins since they form less destructive steric interactions with proteins compared to larger compounds. Furthermore, it has been argued that the size of fragment chemical space is substantially smaller than the chemical space of larger drug-like space such that it can be sampled more efficiently.22 Given these attributes, orders of magnitude fewer molecules are needed within a fragment screening library relative to HTS to achieve “hits”. Fragments tend to bind considerably weaker than larger molecules used in HTS screens (micromolar to millimolar) but with higher ligand efficiency (LE) - average free energy of binding per non-hydrogen heavy atom.22,23 Fragments may be screened against disease targets using a variety of biophysical binding-based techniques that are capable of picking up such weak interactions.21 Once an appropriate fragment hit has been identified, it then serves as a constructive chemical starting point for the generation of a ligand with much greater potency, which may be achieved synthetically by “growing” or “linking” fragments. These fragment optimization strategies are most efficiently executed when a X-ray or NMR structure of the fragment-target complex is available.21,22 In this way, the medicinal chemistry toward optimization is guided in a rational manner requiring fewer analogs to be prepared for delivering a potent lead compound. FBDD is particularly suitable for developing PPI inhibitors, because fragments can access discreet binding pockets on PPI interfaces that larger HTS molecules have not been successful in binding.20,22 Once a suitable fragment anchor is identified to a PPI interface, it can be synthetically elaborated into a molecule making greater opportunistic interactions at the interface capable of perturbing the PPI.
Given their weak interaction with protein targets, fragments typically show little or no activity in enzymatic assays or cell-based assays. Furthermore, many epigenetic proteins possess “reader” functionality by recognition of specific DNA or chromatin marks but are devoid of enzymatic activity making primary screening based on activity challenging. Therefore, biophysical experimental techniques that measures direct fragment-protein binding play a central role in the FBDD paradigm.21,23 Among the biophysical methods, fluorescence thermal-shift assays (TSA), surface plasmon resonance (SPR), nuclear magnetic resonance (NMR) spectroscopy, and X-ray crystallography are the most frequently utilized in FBDD.21 Thermal shift assay , also known as differential scanning fluorimetry (DSF) or ThermoFluor is based on the generalization that the stability of a protein is perturbed by the binding of a ligand. This change in protein stability can be measured by the protein denaturation temperature upon the addition of a putative ligand.24 The TSA is readily amenable to a high throughput format with a fluorescent dye (ex. SYPRO® Orange) that fluoresces when binding to the hydrophobic regions of proteins. In native proteins, the hydrophobic regions are usually buried within protein structures but when the protein is gradually denatured upon heating, the hydrophobic regions become exposed which bind to the added fluorescent dye leading to a temperature-dependent change in fluorescence signal.24 By fitting the fluorescence vs. temperature curve, the transition midpoint for thermal unfolding can be determined (Tm).21,24 TSA is an efficient and economic primary screen, given that a 2000-member fragment library can be screened within a day, and compared to X-ray crystallography and NMR, much less protein is required (~2-3mg protein for screening a 2,500 -member fragment library). However, TSA is usually not used as a stand-alone assay since the Tm shift is often small making hit determination somewhat difficult. Furthermore, TSA tends to have a high false positive/false negative rate.21 Nevertheless, TSA serves as an efficient primary screening method to evaluate a complete fragment library when combined with a high-fidelity secondary biophysical screen (i.e. SPR and NMR) to further evaluate the hits.
NMR spectroscopy is another highly useful technique to screen for fragment-target interactions. There are two types of NMR experiments available: ligand-observed NMR and protein-observed NMR.25,26 For ligand-observed NMR, fragments are added to an unlabeled target protein and the specific NMR pulse sequences are performed to obtain the NMR spectra (hence the term “ligand-observed”) of any fragments bound to the target.25,26 To increase the throughput of ligand-observed NMR experiments, mixtures of 5-10 fragments are typically screened.26 Protein-observed NMR can only be used for smaller proteins (<40kDa), and target protein needs to be labeled by isotopes (one or more of 15N, 13C and 2H) during production [25,26]. If the resonances of protein are assigned, the protein residues that bind to fragments can be identified through changes in chemical shifts in the 2D NMR spectra. Comparatively, ligand-observed NMR requires much less protein than protein-observed NMR and is more economical given that no labeling is required.26 Thus, the current trend is to use ligand-based NMR for primary screen and protein-based NMR as a secondary method to identify the fragment binding pocket on protein to guide lead optimization.
Surface plasmon resonance (SPR) is a biophysical experiment that involves the immobilization of a protein on a special and flowing a potential ligand over the surface.21,27 The change in refractive index of the media near the surface is proportional to the number of molecules binding to immobilized protein.21 The immobilized protein can be repeatedly tested against various fragments and, depending on protein stability, can be used to screen against 300-600 compounds a day. Therefore, compared to X-ray crystallography and NMR, SPR requires much less purified target protein.27 The throughput of SPR is higher than both X-ray crystallography and NMR such that approximately 2000 fragments can easily be screened within a week. More importantly, by fitting the curve of refractive index changes, the association rate constant (kon), the dissociation rate constant (koff), and the affinity (Kd) can all be determined - a feature useful in both primary screen and secondary assay to guide hit optimization.27
Generally, X-ray crystallography is used as a secondary method to help guide medicinal chemistry efforts to optimize fragment hits identified from other primary screens. Obtaining a high-resolution protein-ligand co-crystal structure is crucial for the success of FBDD, unless the protein NMR structure can be obtained, because the visualization of ligand and its binding pocket can enable chemists with critical structural information necessary to synthetically “grow” a fragment into more potent compounds.22,28 Alternatively, X-ray crystallography can also be used as a primary screen by soaking individual members or mixtures of fragments into preformed crystals of the target. This requires robust protein crystals that are amenable to compound soaking, robots to automatically handling multiple protein crystals, facilities to efficiently obtain X-ray diffraction data, and a computational pipeline to efficiently solve the protein-ligand structures.29 While the throughput of screening a fragment library using X-ray crystallography is much lower than other biophysical techniques, the advantage of this strategy is that each hit renders a valuable protein-ligand co-structure that can be readily used for structure-guided medicinal chemistry.21,29 Furthermore, the hits identified from X-ray crystallography can usually be verified by other biophysical assays.30
Each biophysical assay possesses its strength and weaknesses. In many cases, the hits identified using different biophysical assays do not always overlap.31,32 It is often prudent to use several biophysical methods to screen a fragment library against the same target and select hits to follow-up by weighing in the results from different screens.22 However, as discussed above the throughput and protein consumption are often key issues for consideration and thus may not be feasible. Therefore, another strategy is to use a single method as the primary fragment screen and only use the other biophysical assays as secondary methods to validate hits arising from the primary screen.22 There is no set answer for which strategy is better and thus the FBDD workflow should to be tailored to each case based on the issues related to the particular target being screened, resources, equipment, and the time limitation of each specific drug discovery project.
Although it is perceived that fragments usually have low activity in functional assays, many biochemical assays are used as first line screening method for FBDD. For instance, both AlphaScreen® and time-resolved fluorescence energy transfer (TR-FRET) can be used for identifying fragments that block the interaction between two biomolecules including protein, peptide, and DNA.33,34 In the AlphaScreen® assay (PerkinElmer), one biomolecule is usually biotinylated and attached to the streptavidin-coated donor beads while the other biomolecule binds to the acceptor bead via different mechanisms such as His-tag/Ni-NTA and FLAG-tag/anti-FLAG antibody interactions. When the two biomolecules interact, they bring together the donor and acceptor beads.35 Upon laser excitation, the photosensitizer in the donor bead releases singlet/excited oxygen molecules that can travel up to 200 nm in solution, react with the chemiluminescer in the acceptor beads and eventually stimulate the fluorophore’s light emission at 520-620nm.35 The setting for TR-FRET is similar to AlphaScreen® assay, but instead of using donor and acceptor beads, it uses anti-protein tag (ex. His, FLAG) antibodies and streptavidin that are labeled by a pair of fluorophores to recognize the biomolecules that involve in the binding assay.36 The commonly used fluorophore pairs are europium/allophycocyanin and terbium/phycoerythrin. TR-FRET assay has much lower background than the traditional FRET assay because the fluorescence donors such as europium and terbium have longer fluorescence emission lifetime than compounds such as fluorescein. The slight delay of fluorescence reading allows the short-lived background fluorescence to decay and thus clean energy transfer signal can be measured.36 As we will describe later in this article, when using FBDD to target enzymes, it is possible to use enzymatic assays to directly screen for fragment inhibitors at high compound concentration. In fact, it has been reported in many literatures that the fragments deconstructed from known higher molecular weight enzyme inhibitors still retain partial inhibition activities as the parent compounds.37,38
The in silico methods are also used extensively for identifying potential fragment ligands to target proteins from large online compound database such as ZINC.39-44 High-resolution protein structures and preferably co-crystal structures with other known ligands will significantly increase the success rate by this route. Some research groups directly dock the available online fragment libraries onto protein structures40,45 while others, as we will describe in detail bellow, use a fragment-based in silico procedure called anchor-based library tailoring (ALTA) to select subsets of compounds from online compound library such as ZINC library for high throughput docking.39,41,42 For the ALTA procedure, the molecules from online library are decomposed into fragments, screened against target proteins, and the fragment hits will serve as anchors to retrieve molecules from the original library for another run of high throughput docking.39,41,42 The advantage of the ALTA procedure is that the rigid fragments usually dock more accurately than compounds with many rotatable bonds, and the computational cost can be reduced from the smaller subset of retrieved molecules for high throughput docking.41 These in silico fragment screen can be used to generate hits that serve as starting points for obtaining high affinity inhibitors; however, the success of FBDD programs still require either X-ray crystallography or protein-observed NMR to provide more structural information of ligand-protein interactions.
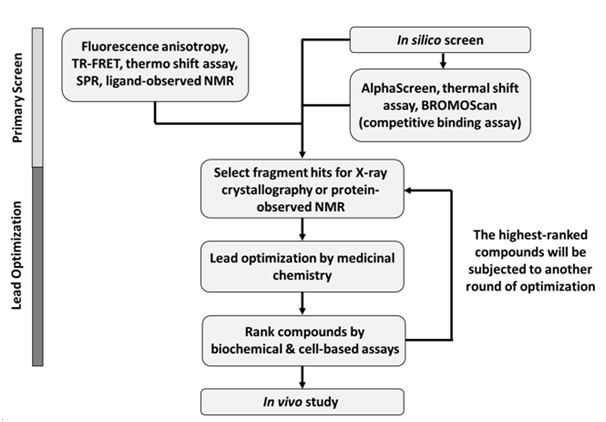
FBDD has been successfully deployed for targeting non-enzymatic epigenetic targets. The most extensive application for FBDD within the epigenetic arena has been toward the bromodomain-containing family of proteins.34,39,41-63 Bromodomains are conserved structural motifs in proteins that recognize acetylated lysine (AcK) on histones.61 There are a total of 61 bromodomains that are found in 42 proteins within the human genome. By recognizing AcK histone marks, bromodomain-containing proteins regulate gene expression either through recruiting other transcription factors or via their catalytic or transcription regulation domains.61 Therefore, compounds that block the interaction between bromodomains and AcK can be used as inhibitors against these proteins. Many bromodomain-containing proteins are implicated in cancer and therefore are interesting drug targets for developing cancer therapeutics.64 One of the major targets in this group is Bromodomain Containing 4 (BRD4). BRD4 contains two N-terminal bromodomains and is essential for c-Myc gene expression in cancer by collaborating with RNA polymerase II to facilitate transcript elongation.65,66 Various FBDD screening methods have been used to find new fragment scaffolds to target BRD4 both in academia and within the pharmaceutical industry. The FBDD strategies for targeting bromodomains are summarized in Figure 1.
In 2012, GlaxoSmithKline assembled a focused set of 1376 fragments containing an AcK-mimetic structure, one or more hydrogen-bounding functionality and a small alkyl substituent.34 The library was screened against BRD4 as well as two other proteins within the same family (BRD2 and BRD3) using a fluorescence anisotropy assay.34 In this assay, they measured the ability of each compound (at 200mM) to displace a fluorescence-labeled benzodiazepine inhibitor from its binding site on BRD2, BRD3, and BRD4. Of the 1367 compounds tested, they identified 132 compounds that show >30% ligand displacement effect against at least one of the three targets.34 To further understand ligand-target interactions, the authors solved at least 40 co-crystal structures of fragments binding to the first bromodomain of BRD2. In a follow-up paper, the same group described optimizing one of the fragment hits into inhibitors of BRD2, BRD3, and BRD4 with Kd of ~500nM.46 Navratilova et al.56 at University of Dundee used SPR to screen a 656 fragment library against the bromodomains of BRD4, PCAF, and CREBBP.56 By screening at fragment concentrations 16.6, 50, and 150mM, they identify hits based on concentration-dependent increased response unit value. Seven fragment hits were selected and the binding affinity (Kd) was determined by SPR at 3-fold concentration series between 1 and 250mM. The authors solved four co-crystal structures of three fragments binding to the bromodomains of BRD4, CREBBP, or PCAF and found that all fragments bind as AcK mimetics and occupy the bromodomain cavity on top of four conserved water molecules in the structure.56
Ligand-based NMR screening was also used to evaluate a library of 539 fragments against the first bromodomain of BRD4 (BRD4(I)).60 The fragments were screened as mixtures of 8-10 compounds at 200mM against 20mM of BRD4(I). The positive hits within each mixture were then confirmed by a second round of ligand-based NMR using individual compound. From the screen, 10 hits were identified and five fragments (at 100mM) were capable of displacing the fluorescence-labeled benzodiazepine inhibitor from BRD4(I) in the fluorescence anisotropy assay.60 Both protein-observed NMR and X-ray crystallography showed that the fragment hits bound to the AcK binding pocket of BRD4(I) and thus could be used as starting points for developing high affinity inhibitors. The TSA assay was also used to screen the fragment library against the bromodomain of PCAF at 1mM and fragments that increase Tm by ~1-2 °C are selected.53 The co-crystal structures of five fragment-bromodomain complexes were solved and compounds with higher affinity (~21mM) to PCAF bromodomain were obtained based on the structure information.53
The uses of in silico screens were deployed by several groups to target the bromodomains of BRD4 and CREBBP with much success.39,41-45,54 Zhao et al.39 at the University of Zurich (Switzerland) used a fragment-based in silico method called Anchor-based Library Tailoring (ALTA) to select compounds from the large (~9 million molecules) ZINC all-now library (version of 2012) for high throughput docking.39 First, the ZINC library was filtered for compounds that are 200-400 Dalton, > 1 hydrogen-bond acceptor, < 8 rotatable bond, and have 2 to 6 rings. This processed library was then automatically decomposed into 375,897 fragments by a published algorithm67 and again filtered to 238,408 fragments against the criteria that each fragment possess a MW between 60-300 Dalton, have ³ 1 hydrogen-bond acceptor, and contain < 4 rotatable bonds.39 This focused fragment library was docked into BRD4(I) and 17,179 fragments were selected as anchor fragments to retrieve 665,184 molecules from the original filtered ZINC library for high throughput docking onto the BRD4(1) structure. From the library, 4826 compounds were predicted to have binding affinity greater than -6.5 kcal/mol (corresponding to 20mM) and a ligand efficiency (LE) more favorable than 0.3kcal/mol per heavy atom. Based on this analysis, 55 compounds were chosen for molecular dynamic (MD) simulation by predicted LE, chemical diversity, rigidity, novelty, and commercial availability. At the end 24 compounds were selected based on the MD simulation results. Four out of the 24 purchased hits are confirmed by positive Tm shifts in TSA assay and the inhibition of AcK histone peptide binding to BRD4(I) in an AlphaScreen assay.39 This research group also used a similar ALTA strategy to perform an in silico screen against the bromodomain of CREBBP and 17 hits were purchased for verification.42 Two out of the 17 selected hits inhibited the interaction between AcK peptide and CREBBP bromodomain at low mM range. Guided by extensive MD simulations and high resolution X-ray crystallography, they optimized both active fragment hits into nanomolar inhibitors of CREBBP bromodomain.42,58
FBDD has also been used to target epigenetic enzymes. Ferreira de Freitas et al.38 at the University of Toronto screened a commercial library of 2040 fragments (at 1mM) against protein arginine methyltransferase 6 (PRMT6), an enzyme that transfers a methyl group from the cofactor S-adenosyl-L-methionine (SAM) to the terminal guanidino nitrogens of arginine on substrates such as histone H4.38 Instead of using biophysical methods which is the traditional approach taken in FBDD, the fragments were directly screened for biochemical enzymatic inhibition of PRMT6 using a scintillation proximity (SP) assay.38 The SP assay used scintillation counting to detect the PRMT6 catalyzed transfer of the labeled methyl group of tritiated SAM (3H-SAM) to biotinylated histone H4 peptide after capturing the peptide onto streptavidin/scintillant-coated microplate. A total of 14 fragments (hit rate: 0.7%) were confirmed to inhibit PRMT6 activity and their IC50 ranged between 0.3 to 430µM.38 Without synthetic optimization, one of the fragment hits demonstrated specificity to type I PRMTs (PRMT1, 3, 4, 6, 8) when tested against a panel of 30 human methyltransferases. Moreover, the fragment could inhibit the enzymatic activity of PRMT6 in HEK293 cells.
Korczynska et al.40 docked the ZINC fragment library, containing over 600,000 fragment molecules, into the active site of a high-resolution structure of lysine demethylase 4A (KDM4A), an enzyme that uses iron(II), α-ketoglutarate, and oxygen to help convert trimethylated lysine on histones into its demethylated form.40 The final list of test compounds was selected from the top 0.1% ranked fragments using following criteria:
To target Sirtuin 2 (SIRT2), a NAD+ - dependent deacetylase that acts on both histone and non-histone targets, Cui et al.37 synthesized a focused fragment library based on two known inhibitors of Sirtuin family: suramin and nicotinamide.37 Their strategy was based on the fact that the binding sites of suramin and nicotinamide are proximal providing the possibility of linking the two fragment anchors together to create high affinity inhibitors and indeed that is the case.37 The fragments were screened against the enzymatic activity of SIRT2 and its close homologs (SIRT1 and SIRT3). After several rounds of optimization, they obtained a highly specific SIRT2 inhibitor with IC50 of ~50nM which could in a dose-dependent fashion increase acetylated tubulin levels in MCF-7 cells.37 Each of the aforementioned success stories suggests that FBDD is a viable strategy for producing lead-like small molecules within the epigenetic target space.
The successes on FBDD depend heavily on the need to gain structural information of ligand-protein interaction by X-ray crystallography and protein-observed NMR even at very early stage of drug discovery process.22 Clear knowledge of the shape of fragment binding pockets and how protein residues interact with compounds can help chemists decide how to synthetically “grow” a fragment hit into a high affinity ligand.28 FBDD requires close collaboration between biophysicists, chemists, and biologist and within a drug discovery team. The initial fragment hits will usually require multiple rounds of chemical modification to obtain the optimized leads that are active in functional/cell-based assays (Figure 1). After conducting the primary biophysical screens, the biophysicists and chemists together must decide the fragment hits for which to follow-up. Afterwards, the biophysicists will produce protein-ligand binding structures by X-ray or NMR for the chemists to perform rational structure-guided chemical optimization. The newly synthesized compounds will be evaluated by simple biophysical or biochemical assays (ex. TSA, TR-FRET, AlphaScreen) and the highest-ranked compounds will again fed into the X-ray crystallography or NMR pipeline for gathering structural information. After several runs of lead optimization, biologists can then test the optimized compound in various cell-based assays and eventually in animal models. Therefore, creating a collaborative interdisciplinary team is crucial for the success of the FBDD paradigm.
The last decade has brought an explosion of interest in epigenetic regulators as therapeutic targets in cancer development. Since epigenetic modifications on DNA and histones are reversible, there is hope that oncogenic epigenetic marks can be transformed to normal marks to halt the cancer development process. Moreover, proteins that recognize specific oncogenic marks can be inhibited. Yet the major challenge within this area is that many epigenetic regulators are not traditional drug targets and only carry out functions through protein-protein or protein-DNA interactions. Moreover, many epigenetic enzymes also contain various non-enzymatic domains for performing specific functions. Targeting the protein-protein interactions of non-enzymatic domains of epigenetic regulators are intrinsically more challenging, but the development of FBDD and biophysical methods increases the possibility of success. Unlike large, drug-like compounds usually found in HTS libraries, the small chemical fragments can access small and non-obvious binding pockets often seen in protein-protein interfaces. Starting from small and soluble fragments, the final optimized compounds generated can be more efficacious and possess better pharmaceutical properties.
We thank Dr. Michael Corsello and Dr. James Campbell for helpful comments on this manuscript. This work was supported by funding from the Cancer Prevention and Research and Institute of Texas (R1314 and RP16065) and the Welch Foundation (H–Q–0042).
The author declares no conflict of interest.

©2017 Chang, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.