Journal of
eISSN: 2377-4282


Research Article Volume 3 Issue 3
1KCL, Incheon, Korea
2Korea University, Seoul, Korea
3Institute of Nanoproduct Safety Research, Hoseo University, Asan, Korea
4Korea Research Institute of Standards and Science, Daejeon, Korea
5Ecopictures, Seoul, Korea
6Hanyang University, Ansan, Korea
Correspondence: Il Je Yu, Institute of Nanoproduct Safety Research, Hoseo University, Asan, Korea, Tel 82-41-540-9630, Fax 82-41-540-9846
Received: January 26, 2016 | Published: March 29, 2016
Citation: Kim JS, Sung JH, Song KS, Dong MS, Lee JH, et al. (2016) Toxicogenomic Analysis for Livers from Sprague-Daley Rats Following 12-Week Inhalation Exposure to Silver Nanoparticles. J Nanomed Res 3(3): 00058. DOI: 10.15406/jnmr.2016.03.00058
Silver nanoparticles (AgNPs) have been extensively applied to many industrial and biomedical fields due to their antibacterial effect. However, a large number of applications is also lead to health and environmental safety concerns. Up to date, it was well-known that AgNPs induced reactive oxygen species (ROS) production, cytotoxicity, pro-inflammatory effect, DNA damage, cell cycle disturb, necrosis and apoptosis by many researches. Also, several studies have been performed to investigate the microarray test for AgNPs in many cell types. However, no work reports the AgNPs toxicogenomic study in liver cell line and tissue until now. For this reason, we performed to in vivo toxicogenomic study for AgNPs inhalation exposed liver tissue. After 12 weeks inhalation exposure to AgNPs for the Sprague-Daley rats, we carried out silver concentration measurement for liver tissues and toxicogenomic analysis. As a result, we found that silver concentrations in livers were dose-dependently increased in male and female rats. However, a gender-different accumulation of silver in the livers did not observe. In toxicogenomic study, we observed that 109 and 150 genes significantly up- and down regulated by AgNPs inhalation exposure in male and female rats, respectively. The significantly altered male rat genes were involved in 54 biological pathways which were typically related with diabetes and metabolism. In female rat, the significantly expression changed genes were involved in 89 biological pathways which were mainly connected with metabolism and cell signaling. Plus, the gender-dependent gene expression changes of more than 2 fold were linked to 240 genes, with 114 genes in the male livers and 126 genes in the female livers. These were related to steroids and xenobiotics metabolism pathway.
Keywords: Silver nanoparticles, in vivo toxicogenomic study, Liver tissue, Inhalation, Gene expression
AgNPs, Silver Nanoparticles; ROS, Reactive Oxygen Species; SPF, Specific-Pathogen-Free; OEL, Occupational Exposure Limit; DMAS, Differential Mobility Analyzing System; SMPS, Scanning Mobility Particle Sizer; GMD, Geometric Mean Diameter; GSD, Geometric Standard Deviation
Nanoparticles increasingly used in consumer and industrial products, due to distinctive physicochemical properties as high reactivity, color change, lower melting temperature and greater solar radiation absorption.1 One of the most commonly used nanoparticles is silver nanoparticles (AgNPs) which have been applied to clothing, food industry, paints, and electronics.2-6 Also, AgNPs used in medical applications as the constituent elements of catheters, implant surfaces, dental alloys and for treating wounds and burns-related infections as well as in drug delivery in cancer therapies.7-10 Such an increasingly widespread usage of AgNPs leads to the exposure of humans, animals and plants through industrial or domestic waste which could produce harmful biological response.11 The target organs for silver nanoparticles have been shown to be the liver in a 28-day oral toxicity study12-13 and 90-day oral subchronic study,14 and the liver and lungs in 90-day inhalation studies.15-16 It was reported that AgNPs induced reactive oxygen species (ROS) production, cytotoxicity, pro-inflammatory effect, DNA damage, cell cycle disturb, necrosis and apoptosis in various in vitro test.17-20 However, in vitro studies could not reflect cell-cell and cell-matrix interaction and hormonal effect.21 For this reason, the importance of the in vivo studies has already been highlighted in the nanotoxicology fields.22 Additionally, the exact mode of action for silver nanoparticle was still poorly known. Accordingly, this study conducted a toxicogenomic examination of rat livers following 12 weeks of inhalation exposure to AgNPs. Fresh air exposed controls were compared with low and high concentration AgNPs exposed groups of male and female rats.
Generation of silver nanoparticles
The silver nanoparticles were generated as described in previous reports.23 The rats were exposed in a whole-body-type exposure chamber (1.3m3, Dusturbo, Seoul), which consisted of a small ceramic heater (50 × 5 × 1.5 mm3) that was housed within a quartz tube case (70mm diameter, 140mm length). Connected to an AC power supply, the heater achieved a surface temperature of about 15000C within a local heating area of 5 × 10 mm2 within about 10s. For the long-term testing, the source material (about 160 mg) was positioned at the highest temperature point. Clean (dry and filtered) air was used as the carrier gas, and the gas flow was maintained at 30 L/min (Re=572, laminar flow regime) using a mass flow controller (MFC, AERA, FC-7810CD-4V, Japan).23 In this study, the system produced different concentrations of nanoparticles (high, medium, and low) in three separate chambers. The nanoparticle generator was operated for 45.99±0.02 L/min (mean±SE) using the MFC and mixed with 200 L/min at the main flow rate through the high-concentration chamber. Using the MFC for the first dilutor (27.19±0.05 L/min, mean±SE), a portion of the high-nanoparticle-concentration was then diverted to the medium-concentration chamber. Similarly, a portion of the medium-nanoparticle-concentration was then diverted to the low-concentration chamber using a second MFC (2.33 ± 0.01 L/min).16
Monitoring of inhalation chamber and analysis of silver nanoparticles
In the individual chambers, the nanoparticle distribution with respect to size was measured directly in real time using a differential mobility analyzer (Short type DMA, 4220, HCT Co., Ltd. Korea, range 5–150 nm) and condensation particle counter (CPC, 4312, HCT Co., Ltd. Korea, 0–108/cm3 detection range), in combination referred to as a DMAS (differential mobility analyzing system) or SMPS (scanning mobility particle sizer). Nanoparticles from 4.23 to 46.95 nm were measured using sheath air at 5 L/min and polydispersed aerosol air at 1 L/min, in order to meet the operational conditions for the DMA and CPC, respectively. The particle concentration in the fresh-air control chamber was measured using a particle sensor (4123, HCT Co., Ltd. Korea) that consisted of 2 channels: 300–1000 nm and over 1000 nm.
Animals and conditions
The animal and exposure conditions were as described in the previous report by Song et al.13 We purchased five-week-old specific-pathogen-free (SPF) Sprague Dawley male and female rats from KOATECH Co. (Korea) and acclimated for 1 week before starting the experiments. During the acclimation and experimental periods, the rats were housed in polycarbonate cages (3 rats per cage) in a room with controlled temperature (21.6 ± 1.20C), humidity (43.7 ± 6.8%), and a 12 hr light/dark cycle. The rats were fed a rodent diet (Harlan Teklab, Plaster International Co., Seoul) and filtered water ad libitum. The 6-week-old rats, weighing about 171 g for the males and 135g for the females, were then divided into 4 groups (each group consisted of 17 male rats: 5 rats for 12-week exposure, 4 rats for 4-week recovery, 4 rats for 12-week recovery, and 4 rats for micronucleus test after 12-week exposure; plus 12 female rats: 4 rats for 12-week exposure, 4 rats for 4-week recovery, and 4 rats for 12-week recovery): fresh-air control, low-dose group (target dose, 0.6×106 particles/cm3, 1.0×109 nm2/cm2, 48.76 μg/m3), medium-dose group (target dose, 1.4 × 106 particles/cm3, 2.5 × 109 nm2/cm2, 117.14 381.43 μg/m3) and high-dose group (target dose, 3.0 × 106 particles/cm3, 5.0 × 109 nm2/cm2, 381.43 μg/m3), and exposed to silver nanoparticles for 6 hr/day, 5 days/week, for 12 weeks. The doses were selected based on the current silver dust occupational exposure limit (OEL) (100 μg/m3), the medium dose was similar to the OEL, and the high and low-doses were higher and lower than the OEL, respectively. Eight animals (4 male and 4 female) from the control, low-dose, and high-dose groups were used for the gene expression profile study after 12 weeks of exposure. The animals were examined daily on weekdays for any evidence of exposure-related effects, including respiratory, dermal, behavioral, nasal or genitourinary changes suggestive of irritancy. The body weights were measured at the time of purchase, at the time of grouping, one day after exposure, once a week during the inhalation exposure and recovery, and before necropsy.
RNA preparation and Oligonucleotide chip microarray
After 12 weeks of silver nanoparticle inhalation exposure, the total RNA was isolated from the liver using the TRIZOL reagent (Life Technologies, USA) according to the manufacturer’s instructions. The RNA was then analyzed for quantity and purity based on the absorbance at 260 and 280 nm using NanoDrop ND-1000 (NanoDropTechnologies, Wilmington, DE). The gene expression profiling was performed using the Illumina Rat Ref-12 Expression BeadChip platform that contains 22,226 probes (Illumina Inc., San Diego, CA, USA). The quality control was repeated with labeled cRNA, and the hybridization on the Illumina® Rat Ref-12 Expression BeadChips was only performed with intact samples according to the manufacturers’ recommendations (Illumina® protocol, www.illumina.com). Briefly, 500 ng of total RNA was labeled using an Illumina Total PrepRNA Amplification kit (Illumina Inc., USA) based on cDNA synthesis with an oligo-dT primer containing a T7 RNA polymerase promoter. The double-stranded cDNA was then used in an in vitro transcription reaction, and the generated single-stranded RNA (cRNA) was labeled by incorporating biotin-16-UTP (Roche Diagnosics GmbH, Mannheim, Germany). 750 ng of the biotin-labeled cRNA was hybridized (16 hrs) to a RatRef-12 Expression BeadChip (Illumina, San Diego, CA). The hybridized biotinylated cRNA was then detected with streptavidin-Cy3 (Amersham Biosciences, USA) and quantitated using an Illumina Bead Station 500GX Genetic Analysis System scanner.24,25
Microarray data analysis
Beadstudio v3.0 was used to evaluate the expression signals generated by the Illumina RatRef-12 Expression BeadChip arrays. The gene expression data were input into GenPlexTM v3.0 software (ISTECH Inc., Korea), normalized using the Quantile method, and the normalized data log-transformed using base 2. Thereafter, the fold change and Welch t-test were applied to select the differentially expressed genes (DEGs) using a volcano plot with a fold change threshold of 1.5 fold and p < 0.05 to indicate significance. The 1.5-fold DEGs were clustered based on hierarchical clustering using the Pearson correlation as the similarity measure and complete linkage as the linkage method. In addition, the gene ontology classification was provided by the KEGG database. A three-dimensional principal component analysis (PCA) was used to analyze and visualize the distribution of the 6 different biological sample types (NC, MLK, MHK, FLK, and FHK) within the three orthogonal components, which were linear combinations of the expression profiles of the selected genes. The number of genes used for the PCA was chosen by a Kruskal Wallis H-test, which classified the six different sample types with the maximum accuracy in the error estimation procedure. The weighted K-nearest neighbor (K-NN) classifier and leave-one-out-cross-validation (LOOCV) were used for the unsupervised classification and error estimation, respectively
The generated silver nanoparticles (AgNPs) properties and concentrations were measured regularly in exposure chamber.16 In summary, In the high-concentration chamber, the geometric mean diameter (GMD), geometric standard deviation (GSD), total number concentration, and mass concentration and surface area of the silver nanoparticles were 15.00 ±1.75 nm, 3.24 ± 106 particles/cm3, 381.43 mg/m3 and 4.85 ± 109 nm2/cm3, respectively, in the medium-concentration chamber, these measurements were 14.38 ± 1.64 nm, 1.41 ± 106 particles/cm3, 117.14 mg/m3 and 1.70 ± 109nm2/cm3, respectively, and in the low-concentration chamber they were 14.54 ± 1.62 nm, 0.66 ± 106 particles/cm3, 48.76 mg/m3, and 0.76 ± 109 nm2/cm3, respectively. The concentrations of silver nanoparticles in each chamber were well maintained during the 12-week exposure period (Supplement 1). These data were summarized in Supplement 1. In each exposure chamber, the AgNPs were well maintained during the 12 weeks exposure period. The AgNPs observed by FE-TEM were spherical in shape and non-aggregated/non-agglomerated forms with diameters under 47 nm.16 The diameters were log normally distributed from 4 to 47 nm and the Count media diameter (CMD) and geometric standard deviation (GSD) were 14.09 nm and 1.71, respectively, with a good correspondence to the mobility diameters.16 To investigate tissue distribution of AgNPs, we measured silver concentration in livers. As a result, we observed that the silver concentration in the livers dose-dependently increased in both male and female rats after 12 weeks of inhalation exposure to AgNPs (supplement 2). However, we could not observe the significant gender difference of silver concentration in livers. Then, we analyzed in vivo gene expression profiles with obtained liver tissue after 12 weeks AgNPs inhalation exposure. Using the DNA microarray, the gene expression changes were evaluated for the livers which obtained from male and female rats exposed to the low and high concentration of AgNPs. The over 1.3-fold up- or down-regulated genes (p<0.05) were regarded as significant and used for the data mining categories. Among 403 genes, 109 and 150 genes were observed expression changes by AgNPs inhalation exposure in male and female rats, respectively. (Figure 1A) (Table 1&2).
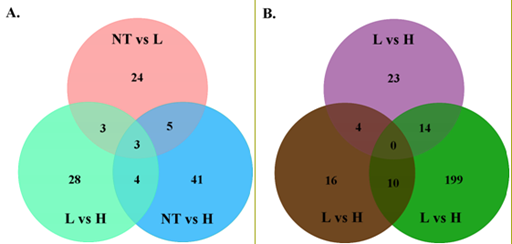
Figure 1 Venn diagram for the numbers of genes expressed significantly different to each control group (Fold change > 1.3, p<0.05).
NT: No Treatment; L: Low; H: High
.
NCBI Accession No. |
Gene Symbol |
Average Fold Change |
||
NT vs L |
NT vs H |
L vs H |
||
NM_053299.1 |
Ubd |
0.183 |
0.47 |
3.612 |
XR_009519.1 |
RGD1562652_predicted |
0.259 |
0.661 |
2.585 |
NM_001008857.1 |
RT1-S2 |
0.379 |
0.664 |
1.754 |
XM_574280.1 |
LOC498989 |
0.479 |
||
NM_001009651.1 |
Clic2 |
0.511 |
1.777 |
|
XM_001063078.1 |
Pex11b |
0.564 |
||
NM_024385.1 |
Hhex |
0.588 |
||
XM_575533.1 |
LOC500181 |
0.638 |
1.717 |
|
XM_001061818.1 |
RGD1565561_predicted |
0.652 |
0.676 |
|
XM_228301.4 |
RGD1311375_predicted |
0.654 |
||
XM_001063114.1 |
Prcp_predicted |
0.664 |
||
NM_207603.1 |
Fcgr3a |
0.685 |
0.74 |
|
XM_001068101.1 |
isg12(b) |
0.699 |
0.691 |
|
NM_019316.1 |
Mafb |
0.705 |
||
XM_578320.1 |
LOC502820 |
0.706 |
0.743 |
|
NM_198740.1 |
Hla-dmb |
0.725 |
||
XM_001066853.1 |
RGD1562052_predicted |
0.733 |
||
NM_012896.1 |
Adora3 |
0.738 |
||
XM_232259.3 |
LOC312688 |
0.752 |
0.654 |
|
XM_001056311.1 |
Fhl3_predicted |
0.754 |
||
XM_001061010.1 |
RGD1564163_predicted |
0.755 |
||
NM_001025750.1 |
Plek |
0.762 |
1.408 |
|
NM_001024289.1 |
LOC499300 |
0.765 |
||
NM_053535.1 |
Enpp1 |
0.768 |
||
XM_231833.4 |
Ppm1k_predicted |
1.302 |
||
XM_001053727.1 |
Bmp7 |
1.374 |
||
NM_053371.1 |
Fxc1 |
1.382 |
||
NM_001012072.1 |
Ppp1r3c |
1.393 |
||
XM_575738.1 |
LOC500380 |
1.394 |
||
XM_576498.1 |
LOC501086 |
1.399 |
||
XM_574174.2 |
RGD1562987_predicted |
1.403 |
||
NM_022701.1 |
Flot1 |
1.405 |
||
NM_057133.1 |
Nr0b2 |
1.481 |
||
NM_013098.1 |
G6pc |
1.52 |
||
XM_579222.1 |
RT1-149 |
1.522 |
||
NM_001024334.2 |
LOC500300 |
1.79 |
2.056 |
|
NM_013144.1 |
Igfbp1 |
2.137 |
||
NM_012703.2 |
Thrsp |
0.526 |
||
NM_173096.2 |
Mx1 |
0.529 |
0.658 |
|
NM_031742.1 |
Kcnh1 |
0.621 |
||
XM_001078678.1 |
RGD1560242_predicted |
0.628 |
||
NM_001011922.1 |
Nedd9 |
0.64 |
||
XM_238366.4 |
RGD1563633_predicted |
0.669 |
||
NM_012621.3 |
Pfkfb1 |
0.681 |
||
NM_001007235.1 |
Itpr1 |
0.698 |
0.746 |
|
NM_022604.2 |
Esm1 |
0.699 |
||
XR_008204.1 |
RGD1565690_predicted |
0.705 |
||
XM_001072449.1 |
RT1-S3 |
0.713 |
||
XM_224824.3 |
LOC306428 |
0.732 |
||
XM_001075137.1 |
Tyki_predicted |
0.734 |
||
XM_579055.1 |
LOC497712 |
0.738 |
||
XM_573861.1 |
LOC364514 |
0.745 |
||
XM_225947.4 |
Trim36_predicted |
0.745 |
||
NM_032082.1 |
Hao2 |
0.748 |
||
NM_001004277.2 |
Lypla3 |
0.75 |
||
XM_001075502.1 |
Ms4a11_predicted |
0.751 |
||
XM_001054526.1 |
Zbp1 |
0.752 |
||
XM_001065014.1 |
RGD1307882_predicted |
0.756 |
||
NM_133593.2 |
Ap3m1 |
0.757 |
||
XM_001056345.1 |
Rtp4_predicted |
0.761 |
||
XM_001059523.1 |
Rarb |
0.762 |
||
NM_032612.2 |
Stat1 |
0.765 |
||
NM_017159.1 |
Hal |
1.302 |
1.542 |
|
NM_001014193.1 |
RGD1359529 |
1.325 |
||
XM_219785.4 |
Gldc_predicted |
1.327 |
||
NM_031345.1 |
Tsc22d3 |
1.345 |
||
NM_001035255.1 |
LOC502603 |
1.352 |
||
XM_223945.4 |
Samd4_predicted |
1.356 |
||
NM_152936.1 |
Spink3 |
1.366 |
||
XM_214172.4 |
Pabpn1 |
1.419 |
||
NM_001012111.1 |
Lpin1 |
1.432 |
1.616 |
|
NM_031108.1 |
Rps9 |
1.437 |
||
XR_009081.1 |
RGD1564649_predicted |
1.446 |
||
NM_017127.1 |
Chka |
1.459 |
||
XM_577408.1 |
LOC501979 |
1.505 |
||
NM_021762.1 |
Tsn |
1.59 |
||
NM_032074.1 |
Irs3 |
1.613 |
||
NM_022238.1 |
Abcb9 |
1.628 |
||
NM_031776.1 |
Gda |
1.661 |
||
NM_172335.2 |
Gm2a |
1.706 |
||
XM_574013.2 |
LOC498736 |
1.916 |
||
NM_019216.1 |
Gdf15 |
2.002 |
||
NM_212505.1 |
Ier3 |
0.65 |
||
NM_031986.1 |
Sdcbp |
0.695 |
||
NM_133543.2 |
Rdh3 |
0.706 |
||
XM_226988.4 |
Fndc3b_predicted |
0.717 |
||
XM_576619.1 |
LOC501191 |
0.725 |
||
NM_001004261.1 |
RGD1303232 |
0.73 |
||
XM_573464.1 |
LOC498241 |
0.743 |
||
NM_001008859.1 |
Mrps10 |
0.745 |
||
XM_001056150.1 |
LOC362068 |
0.754 |
||
NM_031510.1 |
Idh1 |
0.754 |
||
NM_001012007.1 |
Irgm |
0.755 |
||
XM_001073260.1 |
RGD1306565_predicted |
0.758 |
||
NM_053995.3 |
Bdh1 |
0.766 |
||
XM_213574.4 |
Polr2h_predicted |
0.767 |
||
NM_001014076.1 |
Nol10 |
0.768 |
||
NM_134415.1 |
Cdk105 |
1.313 |
||
NM_017171.1 |
Prkce |
1.316 |
||
NM_153308.1 |
Grina |
1.374 |
||
XM_344454.2 |
LOC364468 |
1.381 |
||
XM_345140.2 |
Arsb |
1.392 |
||
NM_001003706.1 |
LOC360228 |
1.402 |
||
XM_235156.4 |
Ptprb_predicted |
1.409 |
||
NM_001012206.1 |
Phlda3 |
1.435 |
||
NM_012603.2 |
Myc |
1.521 |
||
XM_579988.1 |
LOC499620 |
1.529 |
||
NM_019278.1 |
Resp18 |
2.524 |
||
XM_001054526.1 |
Zbp1 |
0.752 |
||
Table 1 Gene expression associated with silver nanoparticles (AgNPs) inhalation exposure in male livers
C: Fresh-Air Control; L: Low Concentration; H: High Concentration
Gene assession no. |
Gene symbol |
Average fold change |
||
Nt vs l |
Nt vs h |
L vs h |
||
NM_017272.15 |
Aldh1a7 |
0.026 |
||
NM_019314.1 |
Kcnn2 |
0.544 |
||
XM_342602.3 |
RGD1306721 |
0.576 |
||
XM_574196.1 |
LOC498907 |
0.605 |
0.627 |
|
XM_214618.4 |
Abhd3_predicted |
0.613 |
0.675 |
|
NM_001005530.1 |
Slc16a13 |
0.675 |
||
XM_574750.1 |
LOC365566 |
0.719 |
0.767 |
|
XM_223397.3 |
LOC305332 |
0.728 |
0.742 |
|
NM_031073.2 |
Ntf3 |
0.736 |
||
NM_024135.2 |
Limk2 |
0.738 |
||
NM_152790.2 |
Carhsp1 |
0.744 |
0.719 |
|
XM_220690.2 |
LOC287518 |
0.747 |
||
NM_133543.2 |
Rdh3 |
0.751 |
||
NM_020976.1 |
Tmem27 |
0.754 |
||
NM_057114.1 |
Prdx1 |
0.756 |
||
XM_579805.1 |
LOC498268 |
0.759 |
0.761 |
|
NM_053883.2 |
Dusp6 |
0.764 |
||
XM_217254.4 |
Ifrd2_predicted |
1.303 |
||
NM_178096.2 |
Nrep |
1.307 |
0.701 |
|
XM_231599.4 |
RGD1306697_predicted |
1.335 |
1.368 |
|
XM_001058675.1 |
LOC368158 |
1.336 |
||
NM_001037787.1 |
MGC112727 |
1.338 |
1.480 |
|
XM_222245.4 |
Hps4_predicted |
1.343 |
||
XR_007378.1 |
RGD1563521_predicted |
1.348 |
||
NM_001007671.1 |
Cyb5d2 |
1.353 |
||
XM_578335.1 |
LOC502835 |
1.358 |
||
NM_017016.1 |
Hdc |
1.374 |
||
NM_001037355.1 |
Mettl7a |
1.416 |
1.377 |
|
XM_233551.2 |
LOC313615 |
1.421 |
0.768 |
|
NM_001017496.1 |
LOC498335 |
1.426 |
1.439 |
|
NM_173322.1 |
Pnrc1 |
1.426 |
1.449 |
|
XM_579556.1 |
LOC497689 |
1.438 |
||
XM_001054760.1 |
Rnf125_predicted |
1.452 |
||
NM_013215.1 |
Akr7a3 |
1.465 |
0.708 |
|
XM_213769.4 |
Vps37b_predicted |
1.470 |
||
NM_012603.2 |
Myc |
1.490 |
1.914 |
|
NM_199113.1 |
Popdc2 |
1.491 |
1.400 |
|
NM_001009681.1 |
Oasl1 |
1.507 |
||
XM_576044.2 |
RGD1564008_predicted |
1.685 |
||
NM_012571.1 |
Got1 |
1.800 |
||
NM_053380.1 |
Slc34a2 |
1.887 |
||
NM_206950.1 |
Mig12 |
0.447 |
||
XM_342965.3 |
Arhgef19_predicted |
0.449 |
||
XM_213329.4 |
Srebf1 |
0.495 |
||
NM_012703.2 |
Thrsp |
0.534 |
||
XM_216452.4 |
Dhcr24 |
0.544 |
||
NM_017136.1 |
Sqle |
0.580 |
||
NM_013134.2 |
Hmgcr |
0.597 |
||
XM_343823.2 |
Serpina7 |
0.605 |
||
NM_138863.2 |
Ltb4dh |
0.610 |
0.601 |
|
XM_001059113.1 |
Slc35d2_predicted |
0.615 |
||
NM_130408.1 |
Cyp26a1 |
0.631 |
||
NM_022389.2 |
Dhcr7 |
0.644 |
||
NM_013105.1 |
Cyp3a3 |
0.644 |
0.619 |
|
NM_171992.2 |
Ccnd1 |
0.645 |
||
NM_001014216.1 |
RGD1306658 |
0.665 |
||
NM_144748.1 |
LOC246263 |
0.667 |
||
XM_576589.1 |
Ihh |
0.668 |
||
NM_019339.1 |
Rgs12 |
0.673 |
||
XM_238366.4 |
RGD1563633_predicted |
0.675 |
||
NM_017080.2 |
Hsd11b1 |
0.681 |
||
NM_173144.1 |
Cyp3a1 |
0.681 |
||
NM_175762.2 |
Ldlr |
0.695 |
||
XM_216756.4 |
RGD1310769_predicted |
RGD131 |
0.698 |
|
NM_173293.1 |
Olr59 |
0.706 |
||
NM_199101.1 |
Plekha4 |
0.719 |
||
NM_031649.1 |
Klrg1 |
0.719 |
||
NM_001004214.1 |
Nqo2 |
0.730 |
||
XM_342979.2 |
Pgd |
0.733 |
||
NM_138836.1 |
Prss8 |
0.734 |
||
NM_001033694.1 |
Srebf2 |
0.735 |
||
XM_001078178.1 |
Klb_predicted |
0.739 |
||
XM_221047.4 |
Polg2_predicted |
0.740 |
||
XR_008878.1 |
RGD1560462_predicted |
0.742 |
||
XM_577565.1 |
Gcnt2 |
0.742 |
||
NM_001009399.1 |
Nsdhl |
0.745 |
||
XM_342986.2 |
Tas1r1 |
0.746 |
||
NM_001007672.1 |
Tmem98 |
0.751 |
||
NM_203335.2 |
Vkorc1 |
0.752 |
||
XM_575223.1 |
LOC499880 |
0.752 |
||
NM_138710.1 |
Dab2ip |
0.753 |
||
XM_215423.4 |
Sesn1_predicted |
0.754 |
||
XM_575430.1 |
LOC500080 |
0.757 |
||
XM_001078486.1 |
RGD1308481_predicted |
0.757 |
||
NM_053019.2 |
Avpr1a |
0.758 |
||
NM_012770.1 |
Gucy1b2 |
0.765 |
||
NM_001025277.1 |
RGD1308076 |
1.303 |
||
NM_013086.1 |
Crem |
1.304 |
||
NM_001013193.1 |
Tial1 |
1.306 |
||
XM_001063880.1 |
Dusp8_predicted |
1.311 |
||
NM_001007691.1 |
Prss23 |
1.315 |
1.326 |
|
NM_199086.1 |
Nob1p |
1.323 |
||
XM_001076614.1 |
Chic2_predicted |
1.326 |
||
XM_001078898.1 |
LOC691575 |
1.328 |
||
XM_575373.2 |
RGD1564980_predicted |
1.329 |
||
XM_230961.4 |
RGD1306056_predicted |
1.333 |
1.304 |
|
XM_221724.2 |
Nrip1_predicted |
1.338 |
||
XM_341874.3 |
Isg20l1_predicted |
1.346 |
||
NM_031512.1 |
Il1b |
1.347 |
||
NM_022186.1 |
Nrbf2 |
1.351 |
||
XM_573296.1 |
LOC498090 |
1.352 |
1.320 |
|
NM_022289.2 |
Snx16 |
1.353 |
||
XM_236656.3 |
Tmem7_predicted |
1.357 |
||
NM_031732.1 |
Sult1c1 |
1.379 |
1.654 |
|
NM_001015004.1 |
Vgll4 |
1.396 |
||
XM_574989.2 |
RGD1560263_predicted |
1.421 |
||
XM_218704.4 |
Smox_predicted |
1.423 |
||
NM_031335.2 |
Polr2f |
1.436 |
||
NM_012743.1 |
Foxa2 |
1.453 |
||
NM_017340.1 |
Acox1 |
1.463 |
||
XM_001078182.1 |
Snrp1c_predicted |
1.472 |
||
NM_017192.1 |
Edg5 |
1.475 |
||
NM_181090.2 |
Slc38a2 |
1.476 |
||
NM_212505.1 |
Ier3 |
1.477 |
||
NM_031507.1 |
Egfr |
1.495 |
||
XM_222107.4 |
RGD1559988_predicted |
1.539 |
||
XM_001066533.1 |
Nnmt_predicted |
1.572 |
||
NM_019195.2 |
Cd47 |
1.587 |
||
XM_343472.2 |
Cish |
1.688 |
||
NM_053727.2 |
Nfil3 |
1.753 |
||
NM_199115.2 |
Angptl4 |
1.753 |
||
XM_001063345.1 |
Zfp364_predicted |
2.039 |
||
NM_053551.1 |
Pdk4 |
2.058 |
||
NM_001013187.1 |
Slc25a30 |
2.119 |
||
XM_001053888.1 |
Gadd45g |
2.163 |
2.112 |
|
XM_001076548.1 |
RGD1566118_predicted |
2.209 |
||
XM_342803.3 |
Plekhf2_predicted |
2.234 |
||
NM_198750.2 |
Cry1 |
2.264 |
||
NM_058208.1 |
Socs2 |
2.558 |
||
XM_341791.2 |
LOC361510 |
21.003 |
||
XM_232684.3 |
RGD1309085_predicted |
0.674 |
||
NM_030850.1 |
Bhmt |
0.701 |
||
XM_001077068.1 |
Cfd |
1.305 |
||
XM_341578.3 |
Riok3_predicted |
1.312 |
||
XM_001058249.1 |
LOC680665 |
1.323 |
||
NM_021762.1 |
Tsn |
1.339 |
||
XM_001061682.1 |
Hist1h2bp_predicted |
1.361 |
||
XM_574280.1 |
LOC498989 |
1.362 |
||
NM_017196.2 |
Aif1 |
1.363 |
||
NM_001013048.1 |
Igfbp7 |
1.370 |
||
NM_020103.1 |
Ly6c |
1.373 |
||
XM_001079607.1 |
RGD1560542_predicted |
1.425 |
||
NM_012823.1 |
Anxa3 |
1.450 |
||
XM_577145.1 |
LOC501744 |
1.496 |
||
NM_207603.1 |
Fcgr3a |
1.529 |
||
XM_342602.3 |
RGD1306721 |
1.742 |
||
NM_053372.1 |
Slpi |
1.787 |
||
XM_573284.2 |
Ssg1 |
2.433 |
||
XM_213106.4 |
RGD1566136_predicted |
5.336 |
||
NM_001008829.1 |
RT1-A2 |
19.684 |
||
Table 2 Amplitude in mill volts of the Lead-1 of electrocardiography in sheep
C: Fresh-Air control; L: Low Concentration; H: High Concentration
In male rats, 35 genes expressions were significantly changed by low concentration exposure, where 12 genes up-regulated and 23 genes down-regulated (Figure 1 & Table 1). Also, the high concentration AgNPs exposure changed the expression of 53 genes which consisted of 21 up-regulated and 32 down-regulated genes (Figure 1A & Table 1). The 80 genes expression changed by both the low and high concentration exposure, and 8 genes were commonly changed where the only one gene (LOC500300) was down-regulated and 7 genes that increased their expression were Ubd, RGD1562652_predicted, RT1-S2, RGD1565561_ predicted, Fcgr3a, isg12(b) and LOC312688. Among the 109 changed genes by AgNPs exposure, 29 genes were involved in the KEGG pathway and related to 54 biological pathways. We selected 10 representative pathway such as Type I diabetes mellitus, antigen processing and presentation, insulin signaling pathway, cell adhesion molecules, purine metabolism, starch and sucrose metabolism, Type II diabetes mellitus, adipocytokine signaling pathway, TGF-beta signaling pathway and Gap junction (Table 3).
KEGG pathway |
Probe ID |
P-Value |
Type I diabetes mellitus |
RT1-149, RT1-S2, RT1-S3, Hla-dmb |
1.17E-06 |
Antigen processing and presentation |
RT1-149, RT1-S2, RT1-S3, Hla-dmb |
2.81E-06 |
Insulin signaling pathway |
Flot1, Irs3, Ppp1r3c, G6pc |
1.21E-05 |
Cell adhesion molecules (CAMs) |
RT1-149, RT1-S2, RT1-S3, Hla-dmb |
2.32E-05 |
Purine metabolism |
Gda, Enpp1, Polr2h_predicted |
2.61E-04 |
Starch and sucrose metabolism |
Enpp1, G6pc |
0.001037 |
Type II diabetes mellitus |
Irs3, Prkce |
0.001272 |
Adipocytokine signaling pathway |
Irs3, G6pc |
0.003071 |
TGF-beta signaling |
Bmp7, Myc |
0.00336 |
Gap junction |
LOC498736, Itpr1 |
0.005107 |
Table 3 Genes showing expression changes following silver nanoparticles (AgNPs) exposure and their relevant KEGG pathways in male rat livers
In female rats, the low concentration of AgNPs exposure changed the expression of 41 genes, where 24 genes up-regulated and 17 genes down-regulated their expression (Figure 1B & Table 2). Plus, the high concentration of AgNPs exposure changed the expression of 103 genes, where 52 genes up-regulated and 51 genes down-regulated their expression (Figure 1B & Table 2). 131 genes were significantly changed by low and high concentration of AgNP inhalation exposure. Among these genes, 13 genes were commonly up- (7 genes: LOC498335, Mettl7a, Myc, Pnrc1, Popdc2, RGD1306697_predicted, MGC112727) and down-regulated (6 genes: LOC498907, Abhd3_ predicted, LOC365566, LOC305332, Carhsp1, LOC498268). Among the 150 changed genes by AgNPs exposure, 43 genes were involved in the KEGG pathway and related to 89 bioloTable 4gical pathways. Ten representative pathways are described in Table 4.
KEGG pathway |
Probe ID |
P-Value |
Biosynthesis of steroid |
Dhcr7, Hmgcr, Vkorc1, Dhcr24, Nsdhl, Sqle |
8.96E-13 |
Bladder, Colorectal, Endometiral cancer |
Ccnd1, Egfr, Myc |
3.72E-05 |
Fatty acid metabolism |
Cyp3a3, Acos1, Aldh1a7 |
4.81E-05 |
Jak-STAT signaling pathway |
Ccnd1, Cish, Socs2, Myc |
5.54E-05 |
MAPK signaling pathway |
Il1b, Egfr, Dusp6, Myc, Ntf3 |
5.92E-05 |
Cysteine metabolism |
Got1, Sult1c1 |
2.74E-04 |
Gammar-Hexachlorocyclohexane degradation |
Cyp3a3, Sult1c1 |
3.19E-04 |
Histidine metabolism |
Hdc, Aldh1a7 |
5.96E-04 |
Cytokine-cytokine receptor interaction |
LOC498335, Il1b, Egfr |
0.00237 |
Thyroid cancer |
Ccnd1, Myc |
0.001212 |
Table 4 Genes showing expression changes following silver nanoparticles (AgNPs) exposure and their relevant KEGG pathways in female rat livers
The genes with the most significant change in their expression following silver nanoparticle exposure were identified as involved in biosynthesis of steroids, bladder, colorectal, endometrial cancer, fatty acid metabolism, Jak-STAT signaling pathway, MAPK signaling pathway, cysteine metabolism, gamma-Hexachlorocyclohexane degradation, histidine metabolism, cytokine-cytokine receptor interaction and thyroid cancer. Additionally, we analyzed gender difference in liver gene expression levels. According to our result, 240 genes showed a more than 2-fold changes when the male and female rat liver gene expression level compared after AgNPs inhalation exposure. Among these genes, 114 male genes and 126 female genes were increased over 10-fold than their counterparts (Table 5).
Gene association |
Gene symbol |
Fold Change |
P-Value |
XM_001062261.1 |
LOC682605 |
0.0022 |
9.84E-10 |
NM_147212.1 |
LOC259244 |
0.0023 |
1.59E-08 |
NM_198784.1 |
Mup4 |
0.0026 |
1.47E-08 |
NM_147214.1 |
LOC259246 |
0.0027 |
2.09E-06 |
NM_147214.1 |
LOC259246 |
0.0032 |
2.13E-10 |
NM_153312.2 |
Cyp3a2 |
0.0032 |
8.40E-10 |
NM_203325.1 |
Mup5 |
0.0037 |
8.22E-09 |
NM_147212.1 |
LOC259244 |
0.0042 |
3.65E-04 |
XM_233029.3 |
LOC298111 |
0.0048 |
6.32E-06 |
XM_001074217.1 |
Ste2 |
0.0052 |
1.07E-06 |
NM_019184.1 |
Cyp2c |
0.0075 |
2.50E-04 |
XM_578458.1 |
LOC502953 |
0.0086 |
0.005275 |
NM_001003409.1 |
LOC298116 |
0.0088 |
6.06E-08 |
NM_001013098.1 |
Dhrs7 |
0.0094 |
2.30E-10 |
XM_575837.1 |
LOC500473 |
0.0106 |
0.001043 |
NM_147215.2 |
Obp3 |
0.012 |
8.66E-08 |
NM_012584.1 |
Hsd3b |
0.0153 |
3.49E-09 |
XM_001056439.1 |
Stac3_predicted |
0.0282 |
4.61E-05 |
NM_031732.1 |
Sult1c1 |
0.0361 |
1.03E-05 |
NM_134380.1 |
Ust5r |
0.0408 |
9.79E-08 |
NM_001014240.2 |
LOC364773 |
0.0423 |
1.41E-05 |
XM_575821.1 |
LOC500457 |
0.0453 |
7.81E-05 |
NM_152936.1 |
Spink3 |
0.0588 |
1.91E-05 |
NM_017061.1 |
Lox |
0.0616 |
9.16E-06 |
NM_145782.1 |
Cyp3a18 |
0.0668 |
1.15E-04 |
NM_019292.3 |
Ca3 |
0.0871 |
1.23E-06 |
NM_053977.1 |
Cdh17 |
0.098 |
0.015137 |
NM_017070.3 |
Srd5a1 |
10.0837 |
1.16E-04 |
XM_216107.3 |
Cpa2_predicted |
12.5769 |
3.25E-04 |
NM_031572.1 |
Cyp2c12 |
24.0966 |
2.45E-04 |
NM_012630.1 |
Prlr |
38.9271 |
3.12E-06 |
NM_017272.15 |
Aldh1a7 |
40.8982 |
0.005545 |
NM_016998.2 |
Cpa1 |
42.0376 |
0.002719 |
NM_022384.1 |
Ascl1 |
84.8827 |
3.28E-04 |
NM_053781.1 |
Akr1b7 |
85.1113 |
7.73E-08 |
XM_235351.3 |
RGD1564515_predicted |
169.3112 |
1.15E-04 |
NM_022258.2 |
A1bg |
226.3963 |
2.49E-06 |
Table 5 Significant gene changes (over 10 fold) in liver mRNA levels between male and female rats
Among the 240 genes which have over 2-fold changes, 68 genes were involved in the KEGG pathway and related to 79 biological pathways. We sorted the major 10 pathway as Biosynthesis of steroid, polysaturated fatty acid biosythesis, Bile acid biosynthesis, PPAR signaling pathway, Terpenoid biosynthesis, Cytokine-cytokine receptor interaction, Linoleic acid metabolism, Jak-STAT signaling pathway and ABC transporters (Table 6).
KEGG pathway |
Gene counts |
P-Value |
Biosynthesis of steroids |
9 |
0 |
Polyunsaturated fatty acid biosynthesis |
3 |
1.72E-06 |
Bile acid biosynthesis |
3 |
5.36E-06 |
PPAR signaling pathway |
4 |
5.44E-06 |
Terpenoid biosynthesis |
2 |
1.75E-05 |
Cytokine-cytokine receptor interaction |
4 |
9.66E-05 |
Linoleic acid metabolism |
2 |
7.87E-04 |
Pyruvate metabolism |
2 |
8.55E-04 |
Jak-STAT signaling pathway |
3 |
8.88E-04 |
ABC transporters - General |
2 |
0.001233 |
Table 6 Amplitude in mill volts of the Lead-1 of electrocardiography in sheep
It was well-known that Silver nanoparticles (AgNPs) have antimicrobial properties and extensively used in various medical and general applications.2 Thus, consumers and workers may be exposed to AgNPs, which may pose harmful effects on their health.26 Exposure to silver nanoparticles have been shown to induce liver toxicity including increase of cholesterol and alkaline phosphatase and bile duct hyperplasia in 28-day oral toxicity study12-14 and 90-day oral subchronic study14 and a 90-day inhalation studies.15 The toxicity of AgNPs on cells are well established and it was reported that AgNPs induced cytotoxicity in primary liver cells and increased superoxide dismutase and glutathione.27-29 In addition, AgNPs produced reactive oxygen species (ROS) which induced oxidative cell damage and mitochondria-involved apoptosis in human liver cell.2 Moreover, most mechanistic studies were focused on in vitro system than in vivo model.30 To find the exact mode of action, this study conducted a toxicogenomic examination with rat livers which were inhalation exposure for 12 weeks to AgNPs. Nanoparticles exposure could occur via inhalation, dermal contact and ingestion during manufacturing and consuming. So, researchers were administered AgNPs with various ways and then they observed tissue distribution for AgNPs to investigate the toxic effect and target organ.
Especially, liver is one of the most importance target organs. Because, the liver is the first line of defense against for exogenous xenobiotic materials and involved in detoxification of heavy metals, including silver.10,20 Additionally, Kupffer cells, reticulo-endothelial system in the liver, can efficiently remove impurities from the blood.20 Also, many studies reported that AgNPs deposition was observed in the liver after oral, inhalation, intraperitoneal and intravenous administration.20,31 In our study, we also observed AgNPs deposition was observed in liver after 12 weeks inhalation exposure. We found that liver tissue silver contents were dose-dependently increased in both male and female rat after 13 weeks inhalation exposure. However, we could not observe sex difference in silver contents of liver tissues. In case of gold nanoparticles (AuNPs) intravenous injection, AuNPs were rapidly and consistently accumulated in liver and spleen among 25 organs after administration. Also, these liver and spleen tissues were significantly altered to genes related to detoxification, lipid metabolism, cell cycle, defense response and circadian rhythm.32 To elucidate the exact mode of action of AgNPs, we have conducted a toxicogenomic study to rat livers which have been exposed silver nanoparticles for 12 weeks inhalation. 1.3 fold up or down regulated genes (p < 0.05) were regarded as significant and used for data mining. According to Park et al., ICR male mice exposed Ag NPs up to 500 μg/kg by single instillation were conducted for microarrary with the mouse lung tissue at day 1 after exposure.30 Inflammation and tissue damage relative genes were identified as significant gene alteration. Also, more than 1000 genes were up-regulated by 1.3 μg/ml Ag NPs in the human lung epithelial cell line (A549).
The up-regulated genes included member of metallothionein, heat shock protein and histone family.33 In present study, we observed that 109 and 150 genes significantly up- and down regulated by AgNPs inhalation exposure in male and female rats, respectively (Table 3&5). The significantly altered male rat genes were involved in 54 biological pathways which were typically related with diabetes and metabolism. In female rat, the significantly expression changed genes were involved in 89 biological pathways which were mainly connected with metabolism and cell signaling. Furthermore, the gender-dependent gene expression changes of more than 2 fold were linked to 240 genes, with 114 genes in the male livers and 126 genes in the female livers. 68 genes of these genes were involved in the KEGG pathway and related to 79 biological pathways which were related to steroids and xenobiotics metabolism. For this result, liver metabolic enzymes activation was different between male and female rat by AgNPs inhalation exposure. Although, we could not observe the significant gene alterations such as the redox system, inflammation, cell cycle, and apoptosis-related genes, metabolism pathway related genes showed a significant alteration.
Silver concentrations in livers were dose-dependently increased in male and female rats. The used-exposure concentration and tissue accumulated concentration of Ag NPs could not induce pivotal toxicogenomic alterations and did not cause serious toxicity in this study. 109 and 150 genes significantly up- and down regulated by AgNPs inhalation exposure in male and female rats, respectively. The significantly altered male rat genes were involved in 54 biological pathways which were typically related with diabetes and metabolism. In female rat, the significantly expression changed genes were involved in 89 biological pathways which were mainly connected with metabolism and cell signaling. Plus, the gender-dependent gene expression changes of more than 2 fold were linked to 240 genes, with 114 genes in the male livers and 126 genes in the female livers. To surely verify the toxicogenomic-mechanism for AgNPs inhalation exposure, we need to find a significant exposure level and carry the long-term study for recovery groups.
None.

©2016 , et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.