Journal of
eISSN: 2377-4282


Review Article Volume 2 Issue 4
1Department of Chemistry-Physics, University of Quebec at Trois-Rivieres, Canada
2Department of Physics, Dalhousie University, Canada
Correspondence: Tajmir-Riahi HA, Department of Chemistry-Physics, University of Quebec at Trois-Rivieres, C.P.500, Trois-Rivieres (Quebec), G9A 5H7, Canada, Tel 819-376-5011 (Ext. 3310, Fax 819-376-5084
Received: October 21, 2015 | Published: November 30, 2015
Citation: DOI: 10.15406/jnmr.2015.02.00038
We have reviewed the effects of synthetic polymers on DNA compaction and particle formation. Synthetic polymers such as poly (ethylene glycol) PEG-(PEG-3350, PEG-6000), methoxypoly (ethylene glycol) anthracene (mPEG-anthracene), methoxypoly (ethylene glycol) poly (amidoamine) (mPEG-PAMAM-G3), (mPEG-PAMAM-G4) and poly(amidoamine) (PAMAM-G4) alter DNA structure and dynamic. The spectroscopic results and atomic force microscopic (AFM) were analysed and the effect of synthetic polymer complexation on DNA stability, aggregation, compaction and particle formation are discussed. A comparison of the overall binding constants showed that the order of binding PAMAM-G4>PEG-6000>PEG-3350>mPEG-anthracene> mPEG-PAMAM-G4>mPEG-PAMAM-G3. The morphology and ultrastructure of polymer-DNA adducts showed major DNA compaction and particle formation induced by synthetic polymers. The generated information is useful for the application of synthetic nanoparticles in gene delivery.
Keywords: PEG, Dendrimer, DNA, Particle formation, Spectroscopy, AFM
MB, Mitochondrial diseases; mtDNA, mitochondrial DNA; MRI, magnetic resonance imaging; GTCS, generalized tonic-clonic seizure; HSV, herpes simplex virus; ENMG, electroneuromyography
Synthetic polymers of a specific shape and size are widely used as drug and gene delivery tools in pharmaceutical and nanomedicine biotechnology.1,2 Poly (ethylene glycol) and its derivatives show major applications in gene and drug delivery due to their solubility, nontoxicity and biocompatibility.3-6 Dendrimers, a family of cationic polymers, are promising nonviral tools for gene and drug delivery because of a well-defined molecular shape, controlled chemical structure, high water solubility, large number of chemically versatile surface groups, and unique architecture.7-14 It has been shown that synthetic polymers induce significant changes in DNA solubility and structure under given conditions. PEGylation of synthetic polymers such as dendrimers is shown to reduce toxicity and increase biocompatibility and DNA transfection.15-21 It is well demonstrated that synthetic polymers induce DNA aggregation and particle formation.21-25 Therefore, it was of interest to review and compare the effects of several synthetic polymers on DNA compaction and particle formation that are recently reported.22-24
Here, we compare the bindings of DNA to several synthetic polymers such as PEG-3350, PEG-6000 and mPEG-anthracene, mPEG-PAMA-G3, mPEG-PAMAM-G4 and PAMAM-G4 at physiological conditions. The data obtained from multiple spectroscopic measurements and AFM microscopic images will be analysed and the effects of various synthetic polymers on DNA compaction and particle formation are discussed here.
FTIR spectroscopy: Infrared spectra were recorded on a FTIR spectrometer (Impact 420 model), equipped with DTGS (deuterated triglycine sulfate) detector and KBr beam splitter, using AgBr windows. Spectra were collected after 2h incubation of polymer with the DNA solution and measured. Interferograms were accumulated over the spectral range 4000-600 cm-1 with a nominal resolution of 2 cm-1 and a minimum of 100 scans. The difference spectra [(DNA solution + polymer) - (DNA solution)] were obtained, using a sharp band at 968 (DNA) as internal reference. This band, which is due to sugar C-C stretching vibrations, exhibits no spectral changes (shifting or intensity variations) upon polymer-polynucleotide complexation, and cancelled out upon spectral subtraction.26
CD spectroscopy: The CD spectra of DNA and its polymer adducts were recorded at pH 7.3 with a Jasco J-720 spectropolarimeter. For measurements in the Far-UV region (200-320 nm), a quartz cell with a path length of 0.01 cm was used. Six scans were accumulated at a scan speed of 50 nm per minute, with data being collected at every nm from 200 to 320 nm. Sample temperature was maintained at 25 °C using a Neslab RTE-111 circulating water bath connected to the water-jacketed quartz cuvette. Spectra were corrected for buffer signal and conversion to the Mol CD (Δε) was performed with the Jasco Standard Analysis software.26
UV absorption spectroscopy: The absorption spectra were recorded on a Perkin Elmer Lambda 40 Spectrophotometer with a slit of 2 nm and scan speed of 240 nm min-1. Quartz cuvettes of 1 cm were used. The absorbance assessments were performed at pH 7.3 by keeping the concentration of DNA constant (125 µM), while varying polymer contents (5 to 100 µM). The binding constants of polymer-DNA adducts were calculated as reported.26
It is assumed that the interaction between the ligand L and the substrate S is 1:1; for this reason a single complex SL (1:1) is formed. It was also assumed that the sites (and all the binding sites) are independent and finally the Beer’s law is followed by all species. A wavelength is selected at which the molar absorptivities εS (molar absorptivity of the substrate) and ε11 (molar absorptivity of the complex) are different. Then at total concentration St of the substrate, in the absence of ligand and the light path length is b = 1 cm, the solution absorbance is:
In the presence of ligand at total concentration Lt, the absorbance of a solution containing the same total substrate concentration is:
(where [S] is the concentration of the uncomplexed substrate, [L] the concentration of the uncomplexed ligand and [SL] is the concentration of the complex) which, combined with the mass balance on S and L, gives:
where Δε11 = ε11 - εS - εL (εL molar absorptivity of the ligand). By measuring the solution absorbance against a reference containing ligand at the same total concentration Lt, the measured absorbance becomes:
Combining equation (4) with the stability constant definition K11 = [SL]/[S][L], gives:
where ΔA = A – Ao . From the mass balance expression St = [S] + [SL] we get [S] = St/(1 + K11[L]), which is equation (5), giving equation (6) at the relationship between the observed absorbance change per centimeter and the system variables and parameters.
Equation (6) is the binding isotherm, which shows the hyperbolic dependence on free ligand concentration.
The double-reciprocal form of plotting the rectangular hyperbola
is based on the linearization of equation (6) according to the following equation:
Thus the double reciprocal plot of 1/ΔA versus 1/[L] is linear and the binding constant can be estimated from the following equation:
Fluorescence spectroscopy: Fluorometric experiments were carried out on a Varian Cary Eclipse. Solution of mPEG-anthracene (80 µM) was prepared at 25 ±1 °C. Various solutions of DNA (5 to 100 µM) in 10 mM Tris-HCl (pH 7.4) were also prepared at 25 ±1 °C. The fluorescence spectra were recorded at λexc = 300-350 nm and λem 400-450 nm. The intensity variations at 420 nm was used to calculate the binding constant (K) for mPEG-anthracene-DNA adducts.
Ultrastructure of polymer-DNA adducts by AFM images
DNA compaction, condensation and particle formation were observed in the presence of PEG, mPEG-anthracene, dendrimers and PEGylated dendrimers (Figure 1). PEG-3350 and PEG-6000 sample showed clear evidence of complexation by AFM images (Figure 1 Panel A). However, this was not the case for the mPEG-Anthracene sample where naked DNA strands could be observed on the mica surface (Figure 1, panel A & C). For the PEG 3350 sample, the complexation was not complete with the presence of DNA strands with a beaded appearance (Figure 1 Panel A). For the PEG 6000 sample, the complexation was much more extensive. The surface was covered with “fried-egg” aggregates similar to the ones observed previously for DOTAP-DNA mixtures.26 The PEG 6000 complexes had an average height of 7.4 (0.2 nm (n =904) and an average volume of 390000 ± 6600 nm3 (n = 904).
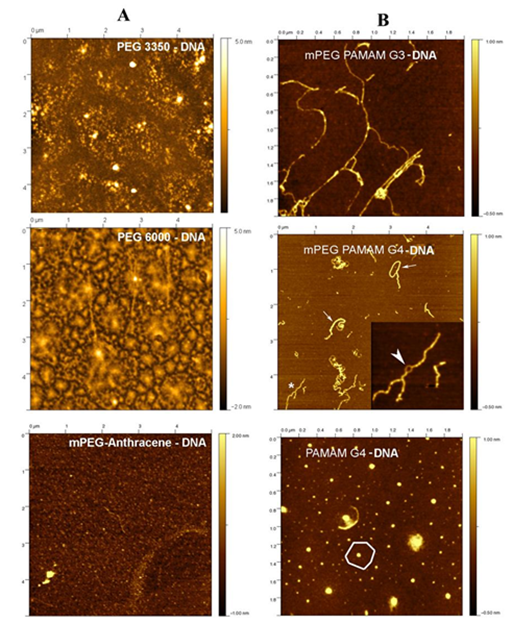
Figure 1 Tapping mode AFM images in air of synthetic polymer-DNA complexes diluted 10 or 100 times in ultrapure water and adsorbed to mica. In all three cases, the surface was covered with aggregates.
Panel A: complexes with PEG 3350, PEG 6000 and mPEG-anthracene and
panel B: for complexes with mPEG-PAMAM-G3, mPEG-PAMAM-G4 and PAMAM-G4.
The AFM images reveal two different types of interactions between the dendrimers and DNA molecules (Figure 1, panel B). The two mPEG terminated dendrimers tend to coat and bundle DNA molecules (Figure 1, panel B). In the case of mPEG-PAMAM-G4, ring-like structures were observed along some of the bundles (Figure 1, panel B) that can be attributed to two “naked” DNA molecules repelling each other (Figure 1, panel B). In contrast, PAMAM-G4 was able to compact DNA into aggregatFigure 1, panel Bes exhibiting a central core surrounded by a flat region (). These complexes were similar to those observed for a mixture of positively charged lipid (DOTAP) and DNA, in a previous study.26 The condensation and compaction of DNA by dendrimers was observed, particularly with PAMAM-G4 (Figure 1, panel B). It is well demonstrated that dendrimers induce DNA compaction and particle formation.22,24
Binding process of synthetic polymers to DNA duplex
Synthetic polymer complexes with DNA via hydrophilic, hydrophobic contacts, groove binding and phosphate interaction.27-31 The infrared spectra and difference spectra of the free DNA showed major alterations of DNA in-plane vibrations at 1710 (guanine), 1661 (thymine), 1610 (adenine) and the backbone phosphate at 1225 asymmetric (PO2) and 1088 cm-1 symmetric (PO2) stretching bands.27-31 upon polymer complexation (Figure 2, panels A&B). Low concentration (0.125 mM) of synthetic polymers PEG-3350, PEG-6000, mPEG-PAMAM-G3, mPEG-PAMAM-G4 and PAMAM-G4 induced minor changes of DNA vibrational frequencies, while at high polymer content (1 mM) major alterations of DNA in-plane and the backbone vibrational frequencies (Figure 2, panels A&B). The major intensity increases were associated with the guanine at 1710 (guanine N7), thymine at 1661 (thymine O2) and adenine at 1610 cm-1 (adenine N7) in the difference spectra of PEG-3350, PEG-6000, mPEG-anthracene, mPEG-PAMAM-G3, mPEG-PAMAM-G4 and PAMAM-G4 complexes of DNA (Figure 2, panels A & B, diffs, 1 mM). The observed intensity changes (particularly at high polymer content) were attributed to polymer interactions with DNA guanine N7, thymine O2 and adenine N7 sites.24,25 Similarly, increase in the intensity of the backbone PO2 groups at 1225 (asymmetric PO2) and 1088 cm-1 (symmetric PO2 vibrations) were observed due to synthetic polymer-PO2 interaction (Figure 2, panels A&B, diff., 1 mM).
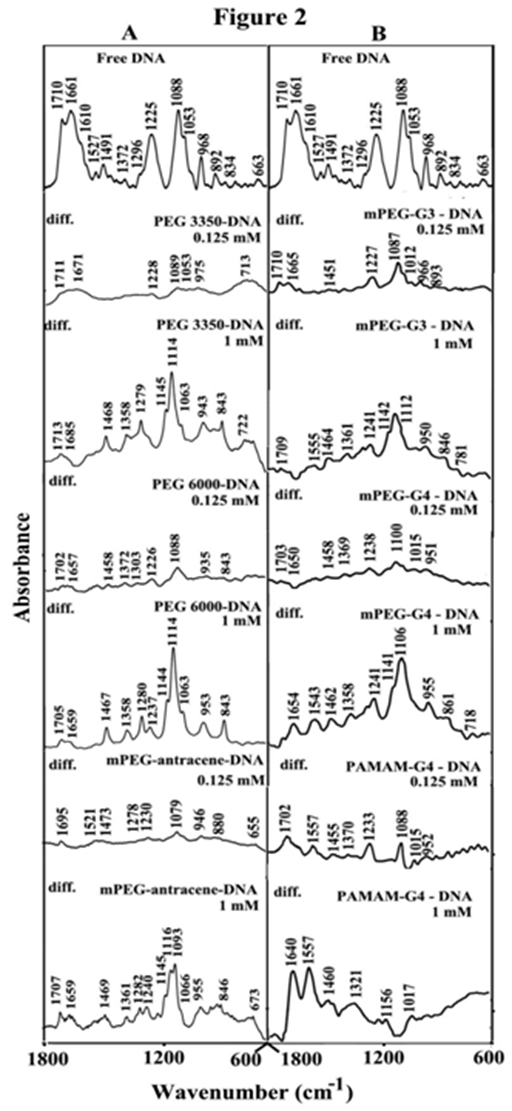
Figure 2 FTIR spectra and difference spectra [(DNA solution + polymer solution) -(DNA solution)] in the region of 1800-600 cm-1 for the free DNA and its synthetic polymer complexes with PEG-3350, PEG-6000 and mPEG-anthracene (panel A) and for mPEG-PAMAM-G3, mPEG-PAMAM-G4 and PAMAM-G4 (panel B) in aqueous solution at pH 7.3 with various polymer concentrations (0. 125 and 1 mM) and constant tRNA content (12.5 mM).
The role of hydrophilic and hydrophobic contacts in polymer-DNA adducts
The shifting of the OH stretching of the free PEG at about 3430 cm-1 to a lower frequency in the infrared spectra of PEG-DNA complexes was attributed to the hydrophilic interaction between PEG and DNA polar groups. Similarly, the shifting of the NH stretching vibration at 3280 cm-1 in the spectra of the free dendrimer and pegylated dendrimerrs was due to the hydrophilic contacts between dendrimer terminal NH2 groups and the DNA polar groups.24,25 However, hydrophobic interactions between DNA and synthetic polymer were characterized by the shifting of the polymer antisymmetric and symmetric CH2 stretching vibrations, in the region of 3000-2800 cm-1. The CH2 bands of the free PEG at 3000, 2990, 2940 cm-1 exhibited a minor shifting, while the CH2 vibrations related to mPEG-PAMAM-G3 located at 2946, 2884 and 2859 cm-1; for free mPEG-PAMAM-G4 at 2942, 2876 and 2856 cm-1 and free PAMAM-G4 at 2969, 2940 and 2834 cm-1 shifted to higher frequencies in the spectra of dendrimer-DNA adducts. The shifting of the polymer antisymmetric and symmetric CH2 stretching vibrations in the region 3000-2800 cm-1 of the infrared spectra suggests the presence of hydrophobic interactions via dendrimer hydrophobic cavities and DNA hydrophobic groups.24,25
Conformation of DNA in polymer complexes
The CD spectrum of the free DNA is composed of four major peaks at 211 (negative), 220 (positive), 245 (negative) and 275 nm (positive) (Figure 3). This is consistent with CD spectra of double helical DNA in B conformation.32,33 As polymer-DNA complexes formed, a major increase in molar ellipticity of the band at 210 nm occurred and the amplitude of the band at 245 was reduced, while the intensity of the band at 275 decreased at high polymer concentration (Figure 3, panel A). However, no major shifting was observed for the band at 275 nm in the spectra of polymer-DNA complexes (Figure 3, panels A & B). This is due to the presence of DNA in B-conformation both in the free state and in the synthetic polymer-DNA adducts. This is also consistent with the infrared results that showed free DNA in B-conformation with IR marker bands at 1710 (G), 1225 (PO2), 892 and 834 cm-1 (ribose-phosphate) with no major shifting of these bands in the polymer-DNA complexes (Figure 2, panels A & B).
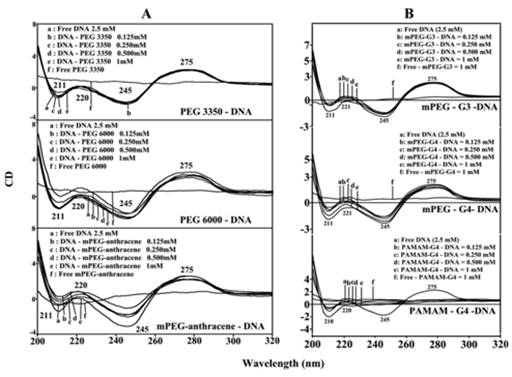
Figure 3 CD spectra of DNA in Tris-HCl (pH ~ 7.3) at 25 °C (2.5 mM) with PEG-3350, PEG-6000 and mPEG-anthracene (panel A) and mPEG-PAMAM-G3, mPEG-PAMAM-G4 and PAMAM-G4 (panel B) with 0.125, 0.25, 0.5 and 1 mM polymer concentrations.
The reduced intensity of the band at 275 nm, in the spectra of polymer-DNA complexes together with the major intensity changes of the band at 210 and 220 nm were attributed to the condensation and particle formation of DNA, in the presence of PEG-3350, PEG-6000, mPEG-anthracene (Figure 3, panel A) and mPEG-PAMAM-G3, mPEG-PAMAM-G4 and particularly in PAMAM-G4-DNA adducts (Figure 3, panel B). The extent of decrease of intensity was much pronounced in the case of PAMAM-G4 nanoparticles, where DNA condensation, compaction and particle formation were observed (Figure 3, panel B). This is consistent with AFM images of the synthetic polymer-DNA complexes that showed major DNA condensation and particle formation by PAMAM-G4 nanoparticles (Figure 1, panel B).
Stability of polymer-DNA adducts
A comparison of the stability of synthetic polymer-DNA adducts by UV-visible spectroscopy .34 showed KPEG 3350-tRNA= 7.9 x 103 M-1, KPEG 6000-tRNA = 1.5 x 104 M-1 and KmPEG-anthracene= 3.6 x 103 M-1 , KmPEG-G3= 1.5 x 103 M-1, KmPEG-G4= 3.4 x 103 M-1 and KPAMAM-G4= 8.2 x 104 M-1 (Figure 4, panels A & B) .23,24 Stronger polymer-DNA complexation formed by PEG-6000 than PEG-3350 and mPEG-anthracene, while PAMAM-G4 forms more stable complexes with DNA than those of PEGylated dendrimers with the order of binding PAMAM-G4>PEG-6000>PEG-3350>mPEG-anthracene>mPEG-PAMAM-G4>mPEG-PAMAM-G3 (Figure 4, panels A & B).24,25 This is indicative of PEG forms stronger complexes than mPEG and PEGylated dendrimers. Similarly, stronger complexes form with larger PEG than smaller PEG. This is also consistent with the conclusion that synthetic polymer-DNA interaction is more hydrophilic than hydrophobic. This conclusion can be supported by the argue that PEG with mostly hydrophilic character forms stronger complexes with DNA, while mPEG-anthracene, with mostly hydrophobic nature forms weaker DNA complexes. Similarly, PAMAM-G4 which has more cationic NH2 groups (64 NH2 groups) than those of mPEG-PAMAM-G4 (32 NH2 groups) and mPEG-PAMAM-G-3 (8 NH2 groups) forms stronger complexes than PEGylated dendrimers (Figure 4, panels A & B).24,25 The results showed that hydrophilic interaction is a major part of synthetic-polymer-DNA complexation.
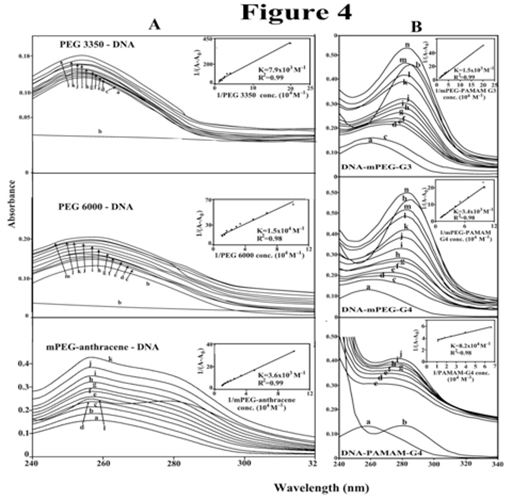
Figure 4 UV-visible results of DNA and its PEG-3350, PEG-6000 (B) and mPEG-anthracene complexes (panel A) and for mPEG-PAMAM-G3, mPEG-PAMAM-G4 and μM PAMAM-G4 complexes (panel B) with free DNA (100 μM); b) free polymer (100); titrated with polymer (5 to 80 μM). Plot of 1/(A-A0) vs (1/polymer concentration) for K calculation of polymer and DNA complexes, where A0 is the initial absorbance of DNA (260 nm) and A is the recorded absorbance (260 nm) at different polymer concentrations (5 μM to 100 μM ) with constant DNA concentration of 100 μM at pH 7.3.
The number of binding sites occupied by polymer on DNA duplex
Since DNA is a weak fluorophore, the titration of mPEG-anthracene was done against various DNA concentrations, using mPEG-anthracene excitation at 330-350 nm and emission at 400-450 nm.35,36 When mPEG-anthracene interacts with DNA, fluorescence may change depending on the impact of such interaction on the mPEG-anthracene conformation or via direct quenching effect.37 The decrease of fluorescence intensity of mPEG-anthracene has been monitored at 420 nm for mPEG-anthracence-DNA systems. The plot of F0 / (F0 – F) vs 1 / [DNA] is shown in Figure 5A. Assuming that the observed changes in fluorescence come from the interaction between mPEG-anthracene and polynucleotides, the quenching constant can be taken as the binding constant of the complex formation. The binding constant obtained was KmPEG-anthracene-DNA = 8.2 x 103 M-1 (Figue 5A’). The association constant calculated for the mPEG-anthracene-DNA adduct suggests low affinity mPEG-anthracene-DNA, which is consistent with the UV results discussed above. The f values obtained in Figure 5, suggest that DNA also interacts with fluorophore via hydrophobic interactions, which is consistent with our infrared spectroscopic results discussed (hydrophilic and hydrophobic contacts).
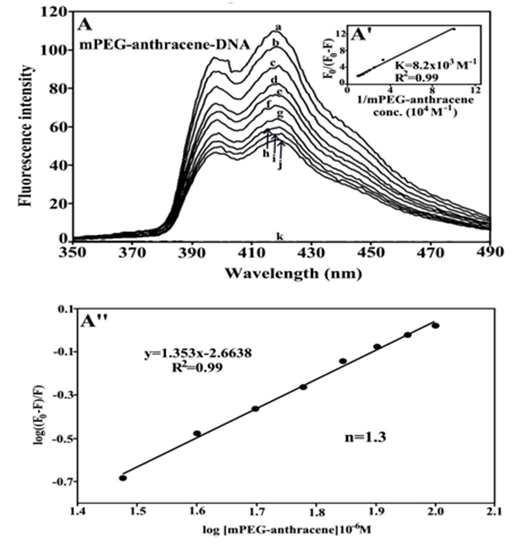
Figure 5 Fluorescence emission spectra of mPEG-anthracene-DNA systems in 10 mM Tris-HCl buffer pH 7.3 at 25 °C for A) polymer-DNA: (a) free mPEG-anthracene (80 μM), (b-j) with polymer-DNA complexes at 5 to 100 μM with (l) free DNA 100 μM. The plot of F0/(F0- F) as a function of 1/DNA concentration. The binding constant K being the ratio of the intercept and the slope for (A’) mPEG-anthracene-DNA. The plot of log (F0-F)/F as a function of log [DNA] for calculation of number of binding sites occupied by mPEG-anthracene molecules on DNA (n) in polymer-DNA adducts (A’’).
The number of binding sites occupied by mPEG-anthracene molecule on DNA (n) was calculated from log [(F0 -F)/F] = logKS + n log [DNA] for the static quenching.38-41 The linear plot of log [(F0-F]/F] as a function of log [DNA] is shown in Figure 5A’’. The n values from the slope of the straight line was 1.3 for mPEG-anthracene-DNA adduct (Figure 5A’’). It seems that about one binding site is occupied by the PEG and mPEG-anthracene on DNA in these polymer-DNA adducts.
Spectroscopic and AFM data of the bindings of several synthetic polymers with DNA were compared here and the following points are concluded. a) Synthetic polymers bind DNA through a major hydrophilic interaction and a minor hydrophobic contact. b) The binding is mainly through polymer polar groups (OH, NH2 and C-O) and DNA bases and the backbone-phosphate group. c) The order of binding is PAMAM-G4>PEG-6000>PEG-3350>mPEG-anthracene>mPEG-PAMAM-G4>mPEG-PAMAM-G3. d) Synthetic polymer complexation induces major DNA condensation, compaction and particle formation, while biopolymer remains in B-family structure. e) This study shows that synthetic polymers have profound effect on DNA morphology that can be of a major importance in gene delivery and DNA transfection. However, major differences were observed between synthetic polymer-DNA complexes and those of the polymer-RNA adducts.42
None.
None.

©2015 , et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.