Journal of
eISSN: 2377-4282


Mini Review Volume 6 Issue 2
Department of Materials and Chemical Engineering, University of Alberta, Canada
Correspondence: Garima Thakur, Department of Materials and Chemical Engineering, University of Alberta, Canada
Received: September 14, 2017 | Published: October 2, 2017
Citation: Thakur G, Thundat T (2017) Designing Protein Self-Assembly for Smart Materials. J Nanomed Res 6(2): 00154. DOI: 10.15406/jnmr.2017.06.00154
Protein self-assembly is advancing in this century as an extraordinary research field. Spontaneous assemblies of proteins or peptides into highly ordered nano-or-microstructures offer unprecedented advances in biotechnology. Designing protein self-assemblies is a challenging field but has huge potential to impact the biosensing industry. In this article we will focus on the most recent chemical and spontaneous self-assemblies of proteins resulting in pattern formation.
Keywords: Self-assembly, Protein patterns, Protein interactions
There is substantial progress made in the field of self-assembly of proteins and peptides in the last two decades.1 Self-assembly of biomolecules into array of nano- or -micro structures facilitate in generation of smart biomaterials.1-3 Biomolecules based nanostructures are extremely sensitive and using proteins as building blocks creates an opportunity to transform the functional micro-environment of the assemblies. Controlled protein self-assembly is still in its infancy but holds great potential in biotechnology.
Protein-protein interactions, essentially subtle non-covalent interactions have major contribution in creating self-assembled structures from protein molecules. Secondary structure of protein molecules such as α-helix and β-sheets serves as a basic unit for the self-assembly. Tertiary structure of protein molecule serves as building blocks and protein-protein interactions (quaternary structures), which may possibly be created using H-bonds and/or Van der Waals interactions initiate and generate the functional self-assembled patterns.1-5 It is important to distinguish between protein and peptide based self-assemblies. Peptide based self-assemblies are largely dependent on secondary structures. Peptides assemble in water at an expense of hydrophobic interfaces formed between building blocks.1 Predominantly two secondary structure elements are used in peptide self-assembly, namely α-helix and β-sheets, nonetheless coils are also used sometimes. Most peptide materials are all α- or all β-materials, mixed assemblies are yet to be reported in our knowledge. It has to be pointed out that it is far easier to modify and control the structure of peptide based assemblies as compared to protein based assemblies. Nonetheless, in this article we will touch upon trends in chemical and spontaneous 1D and 2D assemblies of self-associating proteins to generate complex and diverse patterns. The challenge to design protein assemblies lies in the structure complexity of protein molecules and incomplete understanding of the ways in which they interact to form assembled architectures.5 Here, in this minireview, we have discussed the most recent advances to generate self-associating assemblies of proteins.
One dimensional (1-D) protein self-assemblies
Geometric patterns formed in 1-D include structures such as fibers, and strings. The strategy to design such patterns is based on linking protein monomers in head to tail fashion, which is similar to polymerization process in synthetic chemistry.6 The most salient example is of hemoproteins, such as hemoglobin, myoglobin, cytochromes and peroxidases. Kitagishi and Hayashi have shown linear self-assembly by successive interprotein heme- heme pocket interactions. Here, they have mutated the cyt b562 by replacing His 63 to Cys for site selective modification. Heme modified H63C mutant of cyt b562 undergoes a denaturation at pH 1.9 to remove intrinsic heme, followed by neutralization of aqueous phase to generate heme-modified Apo-Cyt b562 (Figure 1a). AFM image Figure 1b of the pattern formed by H63C on HOPG substrate. Myriad ID self-assemblies are generated using peptides.2 and other biomolecules such as DNA.7 which result in tubular.3 fibrous.1,4 rings.1,4 and other structures, which are not discussed here.
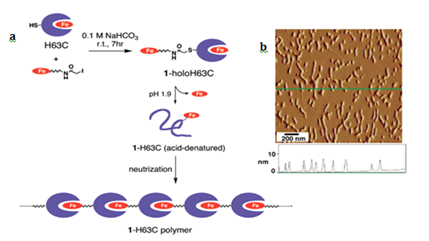
Figure 1 (a) Schematic representation of the supramolecular hemoprotein polymerization. (b) AFM image of the 1a-H63C polymer on the HOPG substrate with a height profile along the green line. Adapted with permission from Reference 5 copyright 2007 American Chemical Society.
Two dimensional protein (2-D) self-assemblies
In field of biosensing, ordered 2-D protein self-assemblies have a great potential. The layered membranes, sheets or ordered structures in 2D offer a huge opportunity in sensing of target molecules and biomolecules as well as in field of biodiagnostics. Two recent distinct examples include computationally designed 2D arrays mediated by non-covalent protein-protein interactions.8 and, metal ion mediated protein assembly under low pH conditions leading to 2D-array.9,10 We have chosen few examples to illustrate the rational designing of protein self-assemblies using different pathways.
Baker et al.8 have used a computational design approach using symmetry docking in Rosetta to self-assemble cyclic protein oligomers into 2D pattern. From protein data bank cyclic oligomer were identified to form six layer groups and based on design calculations most shape-complementary solutions were used to create low energy interface between oligomers. Designs p3Z_42 (Figure 2) from symmetry group P321 with three degrees of freedom, p4Z_9 from group P4212 with three degrees of freedom and lastly design p6_9 from group P6 with two degrees of freedom were generated with cyclic symmetry in lattice. The computationally designed planar protein arrays matched closely with planar 2D crystals generated in vivo and in vitro experiments with size reaching up to 1μm and thickness of 3- 8 nm. Though experiments were not conducted on solid support but the technique set a stage to precisely design 2D protein arrays. However, the challenges in computational approach range from exhaustive sampling and inaccuracies in energy functions leading to larger failure rates.9
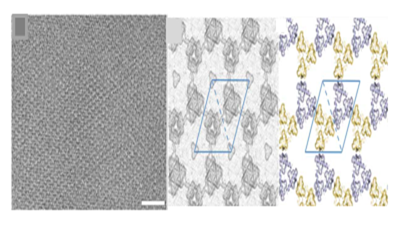
Figure 2 Cryo-EM micrograph of E. coli–grown p3Z_42 and computationally designed models. Reprinted from ref 8 Copyright 2015 American Association for Advancement of Science.
Tezcan et al. [10]devised a strategy with a variant of monomeric heme protein (cytochrome cb562) to form 1D and 2D arrays by changing the pH or concentration of metal ions. cytochrome cb562 was substituted with 3 amino acids to carry Zn+2 and 10 amino acids to promote dimerization into C2 symmetric complex and they termed the variant RIDC3 ( Rosetta Interface Designed Cytochrome 3). The modification of cb562 is similar to the work reported by Hayashi et al. Sizes and morphologies were typically controllable through pH, or ratio of Zn+2 and RIDC3 concentration. The structures converted from microtubules to 2D arrays either at low pH or low [Zn+2]: [RIDC3] ratio (Figure 3). In addition to metal mediated assemblies Suzuki and Tezcan et al..10 have reported the 2D protein lattice of C4 symmetric L-rhamnulose-phosphate-aldoase (RhuA) through disulfide bonds or metal-coordination linkages.11 RhuA variants (Cys modified) self-assembled into 2D open lattice patterns by linking the building blocks. It was remarkable to note that 2D crystal suspensions formed close-packed lattice arrangements at 4 °C when incubated for 1-3 days because of flexibility of single disulfide interactions.
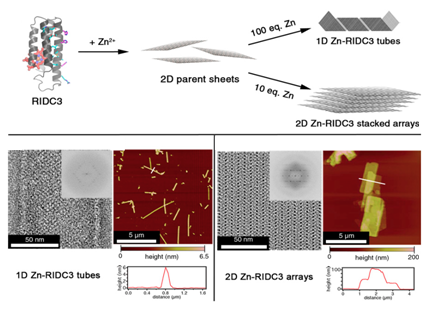
Figure 3 Zn-mediated assembly of RIDC3 into supramolecular arrays and structural characterization of the arrays. Reprinted from reference 7 copyright 2014 National Academy of Sciences.
Other strategies include example of association of two or more proteins to form self-assembled structures, which are not discussed in detail. In one of the examples Gao et al..12 have developed a strategy to self-assemble multiple oligomeric enzymes using protein-protein interactions and enzyme oligomerization. Multienzyme supramolecular device (LPd-FPIMESDs) was created using leucine dehydrogenase (LDH) and formate dehydrogenase (FDH). Octameric LDH was fused with PDZ domain and dimeric FDH was fused with PDZlig at C terminus, which were linked together and 2D organized assembly was generated.
Apart from in-vivo self-assemblies there is a huge attempt to generate controlled 2D protein patterns on substrates. However, for scaffold or substrate based protein pattern designing there are various approaches that are being used such as layer by layer strategy.13 use of protein templates.14 Electrostatic interaction induced assemblies.15 ligand or linker - based protein assemblies.16 gradient-directed assemblies.17 and Langmuir-Blodgett films.18
This mini review highlights some recent chemical and spontaneous protein based self-assembly strategies. Designing of patterns through chemical modification of protein moieties and computational designing and implementation for in vivo and in vitro protein self-assemblies are the two main domains that are touched upon. For biosensing and diagnostics these approaches need to be further developed and implemented. Protein assemblies on substrates present huge opportunity to design patterns using various techniques. However, challenges are faced in designing the real-life applications using self-assembled protein scaffolds. Nonetheless, designing blue prints for biomolecular smart materials with spatial characteristics and active functionalities have vast potential.
No conflict of interest exists.
None.

©2017 Thakur, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.