Journal of
eISSN: 2377-4282


Review Article Volume 3 Issue 5
1Department of Biophysics, Bioinformatics and Computational Omics Lab (BioCOOL), Iran
2Department of Biophysics, Tarbiat Modares University, Iran
Correspondence: Javad Zahiri, Bioinformatics and Computational Omics Lab (BioCOOL), Department of Biophysics, Faculty of Biological Sciences, Tarbiat Modares University, Tehran, Iran
Received: March 04, 2016 | Published: May 20, 2016
Citation: Ramazi S, Zahiri J, Arab SS, Parandian Y (2016) Computational Prediction of Proteins Sumoylation: A Review on the Methods and Databases. J Nanomed Res 3(5): 00068. DOI: 10.15406/jnmr.2016.03.00068
Protein is one of the biological macromolecules, which plays vital roles in the cell. There are numerous post-transitional modifications (PTMs) that strongly affect proteins and their functionality. A PTM occurs when a chemical functional group is being added on or removed from a specific amino acid. A PTM may consist of both enzymatic and non-enzymatic changes. Recent studies have introduced more than 500 different PTM types. SUMOylation is one of the most important PTM; disruption in SUMOylation process affects the cell function and one of the consequences of this change is cell morphology disorder and leads to a variety of sever diseases such as Alzheimer’s disease and Parkinson’s disease. In this paper we have reviewed the current state-of-the-art in silico methods to predict SUMOylation as well as related databases.
Keywords:Post-transitional modification, PTM, SUMOylation, Predicted, Databases, Bioinformatics, Algorithms
Being happened in the nucleus, cytoplasm and organelles of cells; PTM is considered as one of the most important processes in protein functionality .1 Generally, mass spectrometry data are applied to identify the PTMs and their related sites .2 However, the experimental data of PTMs are very limited due to the sophistication and high expenses of the experiments. Recently, the practice of applying the computational methods to predict the protein PTM has interested many researchers .3
Post-translational modification by the Small Ubiquitin-like Modifier (SUMO) proteins, a process termed SUMOylation, is involved in many fundamental cellular processes. SUMOylation is a eukaryotic post-translational modification, which consists of a reversible attachment of members of the Small Ubiquitin-like Modifier (SUMO) protein family on a protein substrate resulting in the dynamic regulation of its biochemical properties .4 Proteins involved in many fundamental cellular processes like DNA repair, transcriptional control, chromatin organization, macromolecular assembly and signal transduction are SUMOylated.5 SUMOylation is also discovered to be involved in various diseases and disorders especially neural ones, such as Alzheimer’s disease and Parkinson’s disease.6-8
The size of SUMO proteins is almost 10 kDa and three-dimensional structures of these proteins are similar to ubiquitin proteins. Interestingly, SUMO proteins have different distribution of surface charge and have less than 20% amino acid sequence similarity.10 During SUMOylation, a SUMO protein is attached to the target protein, which has an acceptor lysine, through three enzymes and then makes the modification. Finally, SUMO is detached by sumo-specific protease.4 (Figure 2). SUMO proteins have been discovered in a wide range of eukaryotic organisms.9 SUMO family has four isoforms in human, one isoform in yeast and eight isoforms in plants .10,11 However, in most vertebrates family SUMO has three isoforms that are known as SUMO1 namely (sentrin، PIC1، GMP1، Ubl1، Smt3c) and SUMO2 namely (sentrin-2, Smt3b) and SUMO3 namely (sentrin-3, Smt3a).11-15 Figure 3 shows the number of papers that have reported experimentally verified SUMOylation in different years, these information are based on the PubMed IDs reported by dbPTM .16 recently published database about PTM data.
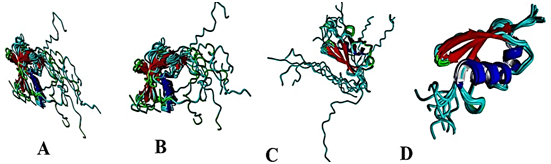
Figure 1 Schematic drawing of the three Sumo proteins using software YASARA.
(A) 2MW5: Solution Structure of Human Small Ubiquitin like Modifier protein-1 (SUMO-1) in Homo sapiens.
(B)1L2N: Smt3 Solution Structure in Saccharomyces cerevisiae. (C) 1WZ0: Solution Structure of Human SUMO-2 (SMT3B), a Ubiquitin-like Protein in Homo sapiens.
(D) 11U4A: Solution Structure of Human Small Ubiquitin like Modifier protein-3 (SUMO-3 C47S) ) in Homo sapiens.
The SUMOylation prediction
Almost computational methods for SUMOylation prediction use sequence information in the neighborhood of the lysine amino acid. Specially considering sequence motifs that are recognized by SUMO Although there are many lysine residues in a protein, but few of these residues in certain motifs are SUMOylation site.17 Many SUMOylation sites contain a consensus sequence motif of WKXE, in which W represents aliphatic amino acids such as I, V, L, A, P or M; X represents each amino acid and E represents glutamic acid. However, the experimental data analysis demonstrates that nearly 23% of SUMOylation sites do not follow SUMOylation of this consensus motif 18,19. In addition to the consensus motif, other SUMOylation motifs are reported such as SUMOylation negatively charged amino acid motif (NDSM: WKXE (D/E), SUMOylation dependent phosphorylation motif (PDSM: WKXEXXSP) and SUMO- Style motif (WKXEP) 20,21 Generally, a computational method uses appropriate features of the potential SUMOylation sites considering the experimentally validated data to train a model for SUMOylation prediction. So, thee availability of the valid databases is crucial to construct accurate models. Then main databases for SUMOylation have been reviewed in Table 1 &2.
|
Database |
A Brief Description |
|
Swiss-Prot/Uniprot database |
This database is one of the largest experimental sources for a variety of post-translational modifications of proteins. (www.ebi.ac.uk/uniprot/) |
|
Tr EMBL database |
This database contains tools and extensive educational tools for both researchers and scholars. |
|
It also provides data about the different types of proteins PTMs and their related changes. |
|
|
DbPTM database |
This DB provides data on post-translational modifications of proteins. Using this database, protein-protein interactions and their specific protein binding positions with the domain could be identified. (http://dbPTM.mbc.nctu.edu.tw/) |
|
HPRD database |
This database is considered as a reference database. It contains data about the human proteins as well as protein PTM data. (http://www.ebi.ac.uk/RESID/) |
|
Phospho site plus database |
Currently, the database contains information on a variety of protein post-translational modifications such as acetylation, methylation, SUMOylation and O- glycosylation. (http://www.phosphorylation.biochem.vt.edu/) |
Table 1 Information on database.
|
Predictor |
Training Feature |
AC |
SN |
SP |
MCC |
AUC |
|
SUMO plot .22 |
Only sequence |
90% |
80% |
93% |
48% |
- |
|
SUMOSP1.19 |
Only sequence |
92.71% |
83.60% |
93.08% |
50.12% |
73% |
|
Boshuliuetal .23 |
Sequence and physicochemical properties |
89.18% |
_ |
_ |
_ |
_ |
|
Find SUMO .24 |
Only sequence |
87.40% |
86.40% |
87.50% |
|
_ |
|
SUMO pre.10 |
Only sequence |
97.71% |
73.96% |
97.67% |
63.64% |
_ |
|
SUMOSP2 .25 |
Sequence and physicochemical properties |
_ |
96.69% |
88.17% |
_ |
_ |
|
SUMO tr .26 |
Sequence and 3D structure and hydrophobicity |
85% |
95% |
75% |
68% |
85% |
|
See SUMO.21 |
Sequence and physicochemical properties |
97.68% |
67.57% |
99.79% |
67.86% |
92% |
|
SUMO hydro .27 |
Sequence and physicochemical properties |
58.30% |
94.40% |
93.30% |
41.90% |
- |
|
SUMO hunt.28 |
Only sequence |
85% |
95% |
75% |
68% |
- |
|
GPS – SUMO.7 |
Only sequence Hydrophobicity |
- |
- |
- |
- |
- |
Table 2Information on predictions SUMOylation articles.
A review of articles from 2004 to 2015, predictions SUMOylation.
Assessing the Performance of the Sumoylation Prediction Methods
There are different assessment measures based the four following basic parameters:
The most important assessment measures based on the above-mentioned parameters have been described in the following.
SS (Formula 1-1)
SS (Formula 1-2)?
AA (Formula 1-3)
MM (Formula 1-4)
SUMOylation is one the most important PTM type, which a disruption in this PTM can lead to various diseases such as type 1 diabetes, Parkinson’s disease, Alzheimer’s disease, heart disease, cancer and brain failure. Considering the cost and limitations of the experimental methods, in the recent years, many studies devoted to computational detection of SUMOylation. In this paper, the databases of experimentally verified SUMOylation and the computational methods for the prediction of SUMOylation have been reviewed. While there are promising methods for the SUMOylation prediction, but considering the limited experimental SUMOylation data, there are considerable rooms to improve the SUMOylation prediction tools.
None.
None.

©2016 Ramazi, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.