Journal of
eISSN: 2373-437X


Research Article Volume 5 Issue 3
1Department of Medical Microbiology, Maternity and Children Hospital in Dammam, Saudi Arabia
2Department of Medical Microbiology, University of Manchester, UK
3Department of Microbiology and Immunology, University of Otago, New Zealand
4School of Biomedical & Healthcare Sciences Plymouth, University Peninsula Schools of Medicine and Dentistry, UK
Correspondence: Mohammed Al Mahrous, Department of Medical Microbiology, Maternity and Children Hospital in Dammam, Saudi Arabia, Tel 96659233472
Received: June 10, 2017 | Published: July 24, 2017
Citation: Mahrous MA, Burnie J, Tagg J, Upton M (2017) Purification and Characterization of 221, the Cationic Peptide that Inhibits Staphylococcus aureus. J Microbiol Exp 5(3): 00152. DOI: 10.15406/jmen.2017.05.00152
Bacteriocin-like inhibitory substances (BLISs) that are produced by various species of Staphylococcus spp., could have potential as topical therapeutic agents for treating highly drug-resistant staphylococcal infections. The spectrum of activity of some such agents, recently identified in our laboratory, has been characterized. This was carried out at different temperatures using deferred antaonism assay. The biological activity of 221, which produced by S. epidermidis strain 221 and shows activity against Epidemic MRSA-15 and strains of MSSA, is described in detail. The initial characterization of this agent showed that its activity is stable on plates pre-exposed to high temperatures (70-80oC/30-40mins) and displaying specificity for S. aureus. This suggests that BLIS 221 is a bacteriocin in nature, possibly of Class-I.
Keywords: drug, antaonism, staphylococcal, antibiotics, proteinaceous, genetic
BLISs, bacteriocin like inhibitory substances
Staphylococcal Resistance to conventional antibiotics used to treat staphylococcal infections has risen to an alarming level.1,2 A contributing factor has been the widespread use of antibiotics to treat non-life-threatening infections. In recent years, there has been much focus on a promising new class of bacteriocins known as lantibiotics.3,4 Bacteriocins are proteinaceous substances produced by certain bacteria, which display growth-inhibitory activity against a range of other bacteria belonging to the same or closely-related species. Bacteriocins of gram-negative bacteria (colisins) have been extensively characterized, resulting in comprehensive knowledge of the genetic basis, domain structure, mode of formation, and killing action of these molecules.5,6 On the other hand, the bacteriocins of gram-positive bacteria have been classified into numerous subtypes.7–9 The first class of bacteriocins (lantibiotics) is distinguishable from other antibiotics because of their polypeptide nature and bioactive properties. We here investigate the administration of the novel cationic peptide antibiotic (221) as a future opportunity for treating staphylococcal infections.
Bacterial strains
S. epidermidis221 was (The University of Manchester, Clinical Microbiology Department Library Strains, Strain no. 221) maintained on Columbia blood agar but was propagated in TSAYca (30g of Oxoid Trypticase soy powder per liter, 20g of Bacto yeast extract per liter, 5g of Bacto Calcium carbonate per liter, 10g of Oxoid Biological agar) at 37oC, then 221 inhibitor was extracted by freeze-thaw method on which 221 strain was grown as a lawn under appropriate incubation conditions. After incubation, the plates were frozen at -70oC for 3-4 hr, and then thawed at room temperature. The frozen agar exudes liquid as it thaws. This was collected, clarified by centrifugation at 15,764g for 5 min and tested for inhibitory activity by the spot or well diffusion assay. Staphylococcal core indicator strains (Table 1) were used in subsequent assays to test for S. aureussusceptibility to activity of 221 strains and used as representative of sensitive, moderate and highly resistant species to synthetic antibiotics. These strains were clinical isolates selected from a wide range of S. aureus library with the exception of an M. leutus strain, which was a reference isolate used commonly as a sensitive biological indicator of BLIS.10
Indicator |
Strain specification |
I1 |
Micrococcus luteus, strain T-181 |
A222 |
Methacillin-sensitive S. aureus strain A222 |
A269 |
Methacillin-sensitive S. aureus strain A269 |
A285 |
Methacillin-sensitive S. aureus strain A285 |
A208 |
Epidemic methacillin-resistant S. aureus type-15 strain A208 |
Table 1 Staphylococcal core indicator strains
1From Tagg and Bannister (1979)
Bacteriocin assay
Bacteriocin production was determined as described previously by either agar well diffusion assay or spot-on-lawn assay.11
221 purification
An overnight culture of S. epidermidis 221 was used to inoculate TSAYca media. The culture was incubated at 37°C, and the cells were then removed using a sterile spatula. The free bacterial cells plate culture was then frozen at -70oC for 3-4h, and then thawed at room temperature. The liquor was then collected and centrifuged at 15,764g for 15min. The liquor was then filter-sterilized by passage through a 0.45-mm-pore-size membrane filter. Solid (NH4)2SO4 was added to 60% saturation at room temperature, and the solution was stirred for approximately 20min until all of the ammonium sulfate had dissolved. A floating pellicle composed of protein and lipid was removed, and the suspension was centrifuged at 15,764g for 15min at 4°C. The supernatant was discarded and the precipitate was dissolved in a minimal volume of 10mM sodium phosphate, (pKa 7.2). After centrifugation at 15,760g for 10min at 4°C to remove insoluble protein, the sample was bound to a cation-exchange column (DIONEX, ProPacTM SCX-10, Analytical, 2×250 mm, Product no. SP5572, Serial no. 0101.) and eluted with an increasing gradient of 1M NaCl at a flow rate of 1ml/min. Buffer A consisted of 10mM (H2NaO4P) (pH 7.2), and buffer B was composed of buffer A and 1M NaCl. Fractions were collected, and analyzed for activity. Fractions containing peak bacteriocin activity were pooled and then passed over a C18 reverse-phase chromatography column (Genesis C18 300 column [part#FM15975E, batch#9532, Jones chromatography]) at flow rates of 1ml/min. Buffer A consisted of 99.9% Millipore water -0.1% trifluoroacetic acid, and buffer B was 99.9% acetonitrile – 0.1% trifluoroacetic acid. Fractions with bacteriocin activity were concentrated by evaporation, and final purification was achieved by a second cycle of C18 reverse phase chromatography with a shallower acetonitrile gradient. The peak fraction of 221 activities was dried by evaporation and resuspended in DH2O, and the total activity and protein content were determined.
SDS-PAGE and bioassay
Analysis of purified 221 preparations was carried out by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).12 After electrophoresis, gels were either silver stained13 or bioassayed by a modification of the method of.14 The gels were soaked in a solution composed of 20% isopropanol and 10% acetic acid for 2h to remove the SDS, followed by rinsing in distilled water for 4h. The washed gels were placed on a BHT-80 agar plate and overlaid with BHT-80 agar (0.7%) containing Micrococcus leutus at a concentration of 108 organisms.ml-1. The plate-gel combination was incubated at 37°C overnight prior to examination for zones of clearing over the bands of 221.
Analytical methods
To precisely determine the size of purified 221, the samples were analyzed on a Lasermat matrix-assisted laser desorption mass spectrometer at the Univeristy of Manchester Biomolecular Analysis Core Facility.
221 production
221 was isolated from the freeze-thaw liquor of cultures prepared in TSAYca medium to minimize the presence of contaminating proteins and peptides. Monitoring of the production of the bacteriocin during the growth of S. epidermidis 221 showed that it was continuously produced, but maximum activity was found in the culture medium at the very end of the exponential and early stationary phase of growth. When production ceased, the bacteriocin that had accumulated maintained its full activity without inactivation. 221 seemed to exist predominantly in an extracellular form. Activity was found mainly in the supernatant fluid (about 96%); the pellet contained about 4% (Figure 1- Figure 3).

Figure 1 Investigating the spectrum of activity of 221 against a range of staphylococcal indicators using deferred antagonism method.
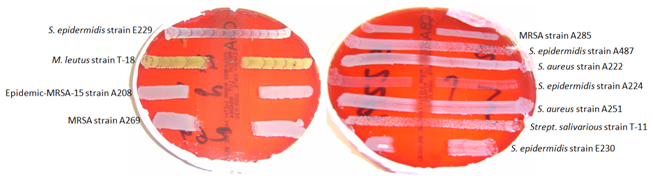
Figure 2 Investigating the spectrum of activity of E221 against a range of staphylococcal indicators using deferred antagonism assay.
Purification of 221
Results of the purification of the bacteriocin from the culture of S. epidermidis 221 are summarized in Table 2. After the last step of purification, 20% of 221 were recovered and the specific activity was _1000-fold higher than that of the culture supernatant. The partially purified peptide was assessed first by Cationic exchange chromatography (Figure 4). Then, the purity of 221 was assessed by HPLC analysis on a C18 column (Figure 5). The elution profile, monitored at 280 nm, demonstrated a distinct absorbency peak which eluted with 56%(v/v) acetonitrile and correlated with the peak of 221 activity. Mass spectrometry analysis indicated that the molecular mass of 221 was between 2000 and 3000 Da.

Figure 5 Separation of the active fractions of 221 from cation-exchange column chromatography by reverse-phase column chromatography.
Purification phase |
Spectrum of activity |
Culture supernatant |
1:16 |
(NH4)2SO4 precipitation |
0.086111 |
CEC1 |
0.397222 |
RPC2 |
0.752778 |
Table 2 Spectrum of activity of the different phases of purification of S. epidermidis strain 221
1Cation-exchange chromatography
2Reverse-phase chromatography
Inhibitory spectrum of 221
By means of the agar diffusion method, all strains of Staphylococcus spp. (including methicillin-resistant Staph. aureus strains), originated from various clinical specimens, were found to be sensitive to purified 221. All these strains were resistant to numerous antibiotics including the third-generation cephalosporin (Cefoxitin).
Lantibiotics are a group of ribosomally synthesized, post-translationally modified peptides containing unusual amino acids. Such amino acids include the thioether amino acids lanthionine (Lan) and/or MeLan, in addition to a number of modified residues, such as 2,3-didehydroalanine (Dha) and 2,3-didehydrobutyrine (Dhb). The presence and influence of these residues on the structure and activity of lantibiotics has been a major hurdle during the sequencing of the 221. This is why Edman degradation is usually necessary for sequencing lantibiotics. In this study, we isolated the bacteriocin-producing strain from a clinical sample. Bacterial characters and laboratory analysis indicated that 221-producing strain was S. epidermidis. The heat-stability and the differences in the spectrum of activity against staphylococcal indicators suggest that 221 is a bacteriocin-type inhibitor. Like most other lantibiotics, 221 is a small cationic peptide and could cause permeabilization of the target cell membrane. The high ammonium sulfate saturation (60%) needed for precipitating 221 suggests that its molecular mass is small. This was confirmed using MALDI TOF/TOF, which seized the mass of 221 in a window mass between 2000 and 3000 Da. The narrow range of inhibitory activity of 221, which was specific for Staphylococcus spp., is also one character of the lantibiotics behaviors.
We thank Dr. Philip Wescombe for stimulating discussions. Many thanks to Dr. David Knight from Manchester Biomolecular Analysis Core Facility for helping in mass-spectrometry. We would like also to thank Marj Howard for her support in operating Ettan-LC. Special thanks also go to Ministry of health in Saudi Arabia for financially supporting the research.
The authors declare that there is no conflict of interest.

©2017 Mahrous, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.