Journal of
eISSN: 2373-437X


Research Article Volume 6 Issue 2
1School of Agriculture and Food Sciences, Jaramogi Oginga Odinga University of Science and Technology, Kenya
2Department of Agriculture and Food Sciences, Meru University, Kenya
3Department of Taita Taveta University, Kenya
Correspondence: Evans Manyara Nyakeri, School of Agriculture and Food Sciences, Jaramogi Oginga Odinga University of Science and Technology, P.O. Box 210-40601, Bondo, Kenya
Received: December 13, 2017 | Published: March 8, 2018
Citation: Nyakeri EM, Mwirichia R, Boga H. Isolation and characterization of enzyme producing bacteria from Lake Magadi, an extreme soda lake in Kenya. J Microbiol Exp. 2018;6(2):57-68. DOI: 10.15406/jmen.2018.06.00189
Microbes inhabitants of extreme environments such as soda lakes are useful in biotechnology as sources of novel enzymes and metabolites, which can survive and function in the harsh conditions of the industrial processes. A number of such extremophiles have been isolated and described from Kenyan lakes. The present study isolated a total of 37 isolates from Lake Magadi, a hypersaline (up to 30% salinity) and alkaline lake (12.5 pH levels), using mineral rich and carbon rich media types. They were characterized and screened for the ability to produce useful biotechnological enzymes and biomolecules by cultural, biochemical and molecular approaches. 34 of the isolates were Gram positive; one was Gram negative and two were Gram variable. Most of the isolates grew well at pH ranging from 6.0–11.0, (optimum 9.0-10.0), temperature range of 20–45oC (optimum of 30-35oC) and salinity range of 5-30%, optimum growth was noted at 10-15%. 15 isolates produced various extracellular enzymes such as amylases, lipases and proteases. Blast analysis of their partial sequences showed that the bacteria belonged to genera Bacillus, Clostridium and Halomonas, with relative abundances of 54%, 38% and 8% respectively. The similarity values of some of the isolates to their closest neighbours (78% 80%, 84%, 85%, 88% and 94-97%) show that they may represent new species or novel genera respectively within the lake ecosystem. The study results are discussed in terms of their ecological importance to the lake system and potential biotechnological application.
Keywords: haloalkalophiles, lake magadi, hypersaline, biotechnology, Bacillus, Clostridium, Halomonas
Micro-organisms that inhabit in environments characterised by conditions which are too harsh for normal life to exist are referred to as extremophiles. Some extremophiles merely tolerate the harsh conditions whereas others require them for their survival and even optimal growth.1 Most described microbial extremophiles are those that thrive under only one distinctive extreme condition such as temperature (thermophiles), salinity (halophiles) or pH (alkaliphiles). Poly extremophiles which have an ability to tolerate and grow under two or more extreme conditions such as haloalkaliphiles, alkalithermophiles and tare rare but in existence.1,2 Haloalkaliphiles, adapted to grow at high salt concentrations and alkaline pH values.3
Soda lakes are alkaline-saline ecosystems that are widely distributed across the globe.4 The unusual geochemistry of these lakes supports the growth of an impressive array of microorganisms that are of ecological and economic importance.5 Lake Magadi (1°43’-2’00’S and 36°13-36o18’E), covering an area of approximately 100 square kilometers is a hypersaline alkaline lake found close to the Kenya-Tanzanian border.
The lake lies at an altitude of 600 metres above sea level, in the rain shadow part of the surrounding mountains.6 The area has two main rain seasons (March-April and August- September) and an average rainfall amount of approximately 500mm per annum.6 With a salinity of up to 35 % w/v or greater and high alkalinity range of 9.0-12, and temperature ranges of between 220C-340C, Lake Magadi s a unique soda lake that has earned itself the tag of an “alkaline saline pan”. This is due to both its large deposits of solid NaCl, Na2CO3 and trona (NaHCO3.Na2CO3.12H2O) and intense rates of up to 3500mm per annum during the dry season.
Though a number of haloalkaliphiles have been isolated and described from this lake,7 its ecology is far from being completely understood or exploited, yet this is nesssary for biotechnological development. In the present study, culture dependent phenotypic characterization and 16S rDNA-based phylogenetic analyses were applied to study aerobic, cultivable bacterial populations present in the alkaline saline lake Magadi. The isolates were further studied for their biotechnological potential. The findings of this study provide information that increases the understanding of the microbial diversity, ecology and potential biotechnological benefits of Lake Magadi microbes, a lake known worldwide as a major source of salt for food and soda ash for glass making.
Sampling
A total of 9 samples were collected from three sampling stations of Lake Magadi in March 2010 and July 2010 namely a hot spring, a salt marsh and a watering point, labelled station 1, 2 and 3 respectively. The samples collected included soda crust, water and sediment (with or without salt). At each sampling site, temperature, pH and total dissolved solutes were recorded (Table 1). Samples from each sampling point were thoroughly mixed to form a representative inoculum for that station.
|
Physicochemical characteristic |
Lonar Lake |
Lake Bogoria |
Lake Nakuru |
Lake Elmenteita |
Mono Lake |
Lake Magadi |
|
pH |
9.8-10 |
10 |
10 |
10 |
9.8 |
9-12.5 |
|
Temperature0C |
26 |
27.9 |
24.1 |
23.1 |
20 |
39.4-51.9 |
|
Salinity % |
1.1 |
4.9 |
1.8 |
2.6 |
8.4 |
30-35% |
|
Dissolved O2(mgL-1) |
13.3 |
13 |
17 |
8 |
3.8 |
NA |
|
Dissolved organic nitrogen (mgL-1) |
53.95 |
2.33 |
11.38 |
8.42 |
NA |
NA |
|
Total phosphorus (mgL-1) |
31.6 |
6.2 |
2.4 |
1.2 |
400 |
NA |
|
Nitrates mgl-1 |
2.4 |
0.3 |
1.4 |
0.3 |
NA |
NA |
|
Cl-( meql-1) |
71.9-87.4 |
176 |
67.3 |
215 |
500 |
6.18 |
|
SO4( meql-1) |
1.0-2.3 |
8.2 |
5 |
14.1 |
260 |
NA |
|
Na+ (meql-1) |
136-159 |
1348.7 |
413.1 |
718.1 |
1300 |
NA |
|
K+ (meql-1) |
0.2-0.4 |
19.2 |
8.5 |
15.9 |
30.7 |
NA |
Table 1 Comparison of physio-chemical parameters of different soda lakes in the world
Source: Table cited in Antony et al.5
Enrichment and isolation of haloalkaliphilic bacteria
Enrichment was done, by inoculating 1 ml of sterile broth media with 0.1 ml of station reperesentative inoculum and incubated overnight at 37°C on a shaking incubator at 200 rpm speed. Serial dilutions of the enriched cells were plated on solid nutrient agar plates prepared using lake water, to replicate the lake conditions of mainly pH and salinity. A total of seven media types were used. Media MI-M4 were carbon rich and adopted from the study of Duckworth et al.,8 while media M5-M8, were mineral rich and adopted from the study of Phil were incubated at 30°C for 5 days. Colonies growing on the plates were selected based on morphological features such as pigmentation and size. Each isolate was subjected to successive streak plating until a pure colony was obtained. Well isolated and differentiated colonies were transferred onto the respective medium slants and cultures and maintained on 20% glycerol stocks (Figure 1) (Figure 2).
Media composition
M1: Glucose 10.0g, Peptone(Difco) 5.0g, Yeast extract(Difco) 5.0g, K2HPO4, 1.Og, MgSO4.7H2O 0.2g, Cycloheximidine 0.01g, Agar 20.0g.
M2: Cellulose 10.0g, Peptone(Difco) 5.0g, Yeast extract(Difco) 5.0g, K2HPO4 1.0 g, MgSO4.7H2O 0.2g, Cycloheximidine 0.01g, Agar 20.0g.
M3: Xylose 10.0g, Peptone (Difco) 5.0g, Yeast extract(Difco) 5.0g, K2HPO4 1.Og, MgSO4.7H2O 0.2g, Agar 20.0g.
M4: Starch 10.0g, Peptone(Difco) 5.0g, Yeast extract(Difco) 5.0g, K2HPO4 1.Og, MgSO4.7H2O 0.2g, Agar 20.0g, Cycloheximidine 0.01g.
M5:(DSC-97): Contained, in grams per litre of Lake Magadi water: Casamino acids, 7.5g, Yeast extract, 10.0g, Trisodium citrate 3.0g, KCl 2.0g, MgSO4.7H2O 20.0g, FeCl2 0.023g, Agar 15g, Cycloheximidine 0.01g.
M6: Cl 1.0 g, MgSO4.7H2O 20.0g, FeSO4.7H2O 0.02g, Trisodium citrate 3.0g, Casamino acids 7.5g, Yeast extract 0.5g, Cycloheximidine 0.01g.9
M8:MgSO4.7H2O 9.7g, Trisodium citrate 3.0g, KCl 2.0g, CaCl2 0.2g, Bacterial peptone(Oxoid L37)10.0g, Agar,15.0g,Cycloheximidine,0.01g.
Morphological and physiological characterization
The isolates were examined for their colony and cell morphology, motility, Gram staining and standard biochemical tests (catalase, oxidase, nitrate reduction, methyl red reaction, acetoin production, citrate utilization by Simmon’s, urease activity, indole production, hydrolysis of casein, gelatin, starch, arginine, carboxyl methyl cellulose (Table 2), growth on nutrient agar with different NaCl concentrations 5%-35% (Table 3), different pH levels (5.5-11.0) (Table 4) and growth at various temperatures (from 5-50°C) (Table 2) according to Bergey’s Manual of Systematic Bacteriology.10
Isolate |
15oC |
20oC |
25oC |
30oC |
35oC |
40oC |
45oC |
50oC |
S2 |
- |
++ |
+++ |
+++ |
+++ |
++ |
+ |
- |
S3 |
- |
+ |
++ |
+++ |
++ |
+ |
- |
- |
S7 |
- |
+ |
+++ |
+++ |
++ |
+ |
+ |
- |
S10 |
- |
+ |
+++ |
+++ |
+++ |
++ |
+ |
- |
S11 |
- |
+ |
+++ |
+++ |
+++ |
++ |
+ |
- |
S12 |
- |
++ |
+++ |
+++ |
++ |
++ |
+ |
- |
X1 |
- |
++ |
++ |
+++ |
+++ |
++ |
+ |
- |
X2 |
- |
++ |
+++ |
+++ |
+++ |
+ |
+ |
- |
X3 |
- |
+ |
+++ |
+++ |
++ |
++ |
+ |
- |
X5 |
- |
+ |
+++ |
+++ |
+++ |
++ |
+ |
- |
X6 |
- |
++ |
+++ |
+++ |
+++ |
+ |
+ |
- |
M8-1 |
- |
+ |
+++ |
+++ |
++ |
+ |
+ |
- |
M8-5 |
- |
+ |
+++ |
+++ |
++ |
++ |
+ |
- |
M8-12 |
- |
+ |
+++ |
+++ |
++ |
++ |
- |
- |
M8-14 |
- |
+ |
+++ |
+++ |
+++ |
+ |
- |
- |
M8-15 |
+ |
++ |
+++ |
+++ |
++ |
+ |
+ |
- |
G2 |
+ |
+ |
+++ |
+++ |
+++ |
++ |
+ |
- |
G4 |
- |
+ |
++ |
+++ |
++ |
++ |
+ |
- |
G8 |
- |
+ |
+++ |
+++ |
+++ |
++ |
+ |
- |
G11 |
- |
+ |
+++ |
+++ |
++ |
++ |
- |
- |
G12 |
- |
+ |
+++ |
+++ |
+++ |
++ |
+ |
+ |
G14 |
- |
+ |
++ |
+++ |
+++ |
+++ |
++ |
- |
G15 |
- |
+ |
+++ |
+++ |
+++ |
++ |
+ |
- |
G18 |
- |
++ |
+++ |
+++ |
++ |
+ |
+ |
- |
G20 |
+ |
+ |
++ |
+++ |
+++ |
++ |
+ |
- |
C1 |
- |
++ |
+++ |
+++ |
+++ |
++ |
+ |
- |
C3 |
- |
+ |
+++ |
+++ |
+++ |
++ |
++ |
- |
C4 |
- |
++ |
+++ |
+++ |
++ |
++ |
+ |
- |
C5 |
- |
+ |
+++ |
+++ |
+++ |
++ |
+ |
- |
C6 |
- |
++ |
+++ |
+++ |
++ |
++ |
+ |
- |
C13 |
- |
++ |
+++ |
+++ |
++ |
+ |
+ |
- |
D2 |
- |
+ |
+++ |
+++ |
++ |
++ |
+ |
- |
D6 |
- |
+ |
++ |
++ |
+++ |
++ |
+ |
- |
D9 |
- |
- |
+++ |
+++ |
+++ |
+- |
+ |
- |
M6-1 |
- |
+ |
++ |
+++ |
++ |
+ |
+ |
- |
M6-7 |
- |
+ |
+++ |
+++ |
+++ |
++ |
+ |
- |
M6-10 |
- |
+ |
++ |
+++ |
++ |
+ |
+ |
- |
Table 2 Tolerance to different temperature levels by the isolates
Key: +growth took ≥5 days; ++ growth took 3-4 days and +++ growth took ≤2 days.
Isolates |
Biochemical tests |
||||||||||||
Arg |
Ly |
Orn |
Gel |
Nitrat |
MR |
VP |
Ca |
Oxid |
Urea |
Cit |
H2S |
Ind |
|
M6-1 |
+ |
- |
+ |
- |
- |
- |
- |
+ |
+ |
+ |
- |
- |
- |
M6-7 |
+ |
- |
+ |
- |
+ |
- |
- |
+ |
+ |
+ |
+ |
+ |
- |
M6-10 |
+ |
- |
- |
- |
- |
- |
- |
+ |
+ |
- |
+ |
+ |
- |
C-1 |
- |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
- |
- |
- |
- |
C-3 |
+ |
- |
+ |
- |
- |
- |
- |
+ |
+ |
- |
- |
+ |
- |
C-4 |
- |
- |
+ |
- |
- |
- |
- |
+ |
+ |
+ |
- |
+ |
- |
C-5 |
- |
- |
+ |
- |
- |
- |
- |
+ |
+ |
+ |
- |
- |
- |
C-6 |
- |
- |
+ |
- |
- |
- |
- |
+ |
+ |
- |
- |
+ |
- |
C-13 |
+ |
- |
+ |
- |
- |
- |
- |
+ |
+ |
+ |
- |
- |
- |
X-1 |
- |
+ |
- |
+ |
- |
+ |
- |
+ |
+ |
- |
- |
- |
- |
X-2 |
+ |
+ |
- |
+ |
- |
+ |
- |
- |
- |
+ |
- |
- |
+ |
X-3 |
+ |
+ |
- |
- |
- |
- |
- |
+ |
+ |
+ |
- |
- |
- |
X-5 |
+ |
- |
+ |
- |
- |
- |
- |
+ |
+ |
- |
- |
- |
+ |
X-6 |
+ |
- |
- |
- |
- |
- |
- |
+ |
+ |
- |
- |
- |
- |
S-2 |
- |
- |
+ |
- |
- |
- |
- |
+ |
+ |
+ |
- |
+ |
+ |
S-3 |
- |
+ |
- |
- |
- |
- |
- |
+ |
+ |
+ |
- |
- |
- |
S-7 |
+ |
+ |
- |
+ |
- |
+ |
- |
- |
- |
- |
- |
- |
+ |
S-10 |
- |
+ |
+ |
- |
+ |
- |
- |
+ |
+ |
+ |
- |
+ |
+ |
S-11 |
+ |
+ |
- |
- |
- |
- |
- |
+ |
+ |
+ |
- |
- |
+ |
S-12 |
- |
+ |
- |
+ |
- |
- |
- |
- |
+ |
- |
- |
- |
- |
D-2 |
+ |
- |
- |
- |
- |
- |
- |
+ |
+ |
+ |
- |
+ |
- |
D-6 |
- |
+ |
+ |
+ |
- |
- |
- |
+ |
+ |
+ |
- |
- |
- |
D-9 |
+ |
+ |
- |
+ |
+ |
+ |
- |
+ |
+ |
- |
+ |
- |
- |
G-2 |
+ |
+ |
+ |
- |
- |
+ |
- |
+ |
+ |
- |
- |
+ |
- |
G-4 |
+ |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
G-8 |
+ |
- |
- |
- |
- |
- |
- |
+ |
+ |
- |
+ |
+ |
- |
G-11 |
- |
- |
+ |
- |
- |
+ |
- |
+ |
+ |
- |
- |
+ |
+ |
G-12 |
- |
- |
- |
- |
- |
- |
- |
+ |
+ |
- |
- |
+ |
- |
G-14 |
- |
+ |
- |
- |
- |
- |
- |
+ |
+ |
- |
- |
+ |
+ |
G-15 |
- |
- |
+ |
+ |
- |
- |
- |
+ |
+ |
- |
- |
- |
- |
G-18 |
+ |
- |
- |
- |
- |
+ |
- |
+ |
- |
+ |
- |
+ |
- |
G-20 |
+ |
- |
+ |
- |
- |
- |
- |
+ |
+ |
+ |
- |
- |
- |
M8-1 |
- |
- |
- |
- |
- |
- |
- |
+ |
+ |
- |
- |
- |
- |
M8-5 |
+ |
- |
- |
- |
- |
- |
- |
+ |
+ |
- |
+ |
+ |
- |
M8-12 |
+ |
+ |
- |
- |
- |
- |
- |
+ |
+ |
+ |
- |
- |
+ |
M8-14 |
+ |
- |
+ |
- |
- |
- |
- |
+ |
- |
- |
+ |
+ |
+ |
M8-15 |
- |
- |
+ |
- |
- |
- |
- |
+ |
+ |
- |
+ |
- |
+ |
Table 3 Biochemical characteristics of the isolates
Isolate |
5.5 |
6 |
6.5 |
7.5 |
8 |
9 |
10 |
11 |
S2 |
- |
++ |
++ |
++ |
+++ |
+++ |
+++ |
+ |
S3 |
- |
++ |
++ |
++ |
+++ |
+++ |
+++ |
+ |
S7 |
- |
+ |
+ |
+ |
+++ |
+++ |
+ |
+ |
S10 |
- |
+ |
+ |
++ |
+++ |
+++ |
+ |
+ |
S11 |
- |
+ |
+ |
+++ |
+++ |
+++ |
++ |
++ |
S12 |
- |
+ |
+ |
++ |
+++ |
+++ |
+++ |
+ |
X1 |
- |
+ |
+ |
+++ |
++++ |
+++ |
++ |
+ |
X2 |
- |
- |
+ |
++ |
+++ |
+++ |
++ |
++ |
X3 |
- |
+ |
+ |
++ |
+++ |
+++ |
++ |
+ |
X5 |
- |
- |
+ |
+ |
+++ |
+++ |
+ |
+ |
X6 |
- |
+ |
+ |
++ |
+++ |
++ |
+++ |
++ |
M8-1 |
- |
+ |
+ |
++ |
+++ |
++ |
++ |
++ |
M8-12 |
- |
+ |
+ |
++ |
+++ |
+++ |
++ |
+ |
M8-5 |
- |
- |
+ |
+ |
+++ |
+++ |
+ |
+ |
M8-14 |
- |
+ |
+ |
++ |
+++ |
+++ |
++ |
+ |
M8-15 |
- |
+ |
+ |
++ |
+++ |
+++ |
+++ |
++ |
G2 |
- |
+ |
+ |
++ |
+++ |
+++ |
+++ |
+ |
G4 |
- |
+ |
+ |
++ |
+++ |
+++ |
++ |
+ |
G8 |
- |
+ |
+ |
++ |
+++ |
+++ |
++ |
+ |
G11 |
- |
++ |
++ |
++ |
++ |
+++ |
++ |
+ |
G12 |
- |
+ |
+ |
++ |
++ |
++ |
+++ |
++ |
G14 |
- |
+ |
+ |
++ |
++ |
+++ |
++ |
+ |
G15 |
- |
++ |
++ |
++ |
++ |
+++ |
++ |
+ |
G18 |
- |
+ |
+ |
++ |
++ |
+++ |
++ |
+ |
G20 |
- |
+ |
+ |
++ |
++ |
++ |
+++ |
+ |
C1 |
- |
+ |
+ |
++ |
+++ |
+++ |
+++ |
+ |
C3 |
- |
+ |
+ |
++ |
++ |
+++ |
++ |
+ |
C4 |
- |
+ |
++ |
++ |
++ |
+++ |
+++ |
+ |
C5 |
- |
+ |
+ |
++ |
++ |
+++ |
++ |
+ |
C6 |
- |
+ |
+ |
++ |
+++ |
+++ |
+ |
+ |
C13 |
- |
+ |
+ |
++ |
+++ |
+++ |
+ |
+ |
D2 |
+ |
+ |
++ |
+++ |
+++ |
+++ |
+ |
+ |
D6 |
- |
+ |
++ |
++ |
+++ |
++ |
+++ |
+ |
D9 |
+ |
+ |
++ |
++ |
+++ |
+++ |
+ |
+ |
M6-1 |
- |
+ |
++ |
++ |
+++ |
+++ |
++ |
+ |
M6-7 |
- |
+ |
++ |
+++ |
+++ |
+++ |
++ |
+ |
M6-10 |
- |
+ |
+ |
++ |
+++ |
+++ |
++ |
+ |
Table 4 Tolerance to different pH levels by the isolates
Key, +growth took ≥5 days; ++ growth took 3-4 days and +++ growth took ≤2 days.
Screening for enzyme production
The ability of the isolates to produce cellulase, xylanase, amylase, lipase, protease and esterase enzymes was investigated by culturing the isolates esparately on basal media (containing per litre of lake Magadi water:1.0g K2HPO4, 0.1g MgSO4.7H2O, 0.05g CaCl2.7H2O and14.0g of Agar), supplemented with 5g of cellulose, 1.25g of xylan., 5.0g of starch, 5ml of olive oil and 1% skim milk respectively (Table 5).11,12 Protease production was suppplemented with the gelatin liquefaction test in which nutrient broth (0.8%) was supplemented with 0.4% gelatin, 1.5% agar, per litre of lake Magadi water and autoclaved. For esterase production, a modified media of Sierra (1957) containing peptone 10.0g, CaCl2.2H2O 0.1g, agar 20.0g per litre of Lake Magadi water and supplemented with previously sterilized 1% (v/v) Tween 80) was used. Each plate of the media was then inoculated with the substrates and incubated at 37oC for 5 days. A clear zone around the colony after 5 days of incubation was taken as evidence of proteolytic activity. The presence of lipase activity was demonstrated by a white halo due to the formation of precipitates of calcium laurate around the growth after 5 days of incubation at 30∘C while the presence of amylolytic activity on the plates was determined by flooding the plate after 5 days with 0.6% KI solution.
Isolates |
Amylase |
Protease |
Lipase |
Cellulase |
Methyl-cellulase |
Xylanase |
Esterase |
G15 |
- |
+ |
- |
- |
- |
- |
- |
C1 |
+ |
+ |
- |
- |
- |
- |
- |
C3 |
+ |
- |
+ |
- |
- |
- |
- |
C4 |
+ |
- |
- |
- |
- |
- |
- |
C13 |
+ |
- |
- |
- |
- |
- |
- |
D2 |
+ |
- |
- |
- |
- |
- |
- |
D6 |
+ |
+ |
- |
- |
- |
- |
- |
D9 |
- |
+ |
- |
- |
- |
- |
- |
M6-10 |
+ |
+ |
+ |
- |
- |
- |
- |
M8-1 |
+ |
- |
- |
- |
- |
- |
- |
M8-12 |
+ |
- |
+ |
- |
- |
- |
- |
S10 |
+ |
- |
+ |
- |
- |
- |
- |
S11 |
+ |
+ |
+ |
- |
- |
- |
- |
S12 |
_ |
+ |
+ |
- |
- |
- |
- |
Table 5 Enzyme producing isolates
DNA Extraction and PCR Conditions
Genomic DNA was extracted from bacterial cells grown aerobically in nutrient broth according to the protocol of Broderick et al.13 1ml of culture cells were suspended in 0.6 ml extraction buffer and incubated at 30oC for 30 minutes. 60µl of 10% SDS and 3µl of 50 mg/ml proteinase K were then added, mixed thoroughly and incubated at 55oC for 2 hours with continuous shaking. 100µl of 5 M NaCl was added followed by thorough mixing and incubation at 720C for 30 minutes. 80µl of CTAB/NaCl mix was added and the mixture incubated at 65oC for 30 minutes to remove cellwall debris, denatured proteins and polysaccharides, after which an equal volume of chloroform isoamyl alcohol (24:1) was added, mixed and centrifuged for 5 minutes, to remove the CTAB complexes. DNA was the extracted by use of phenol-chloroform isoamyl/alcohol (25:24:1) method. The resultant DNA pellet was washed in 1 ml of 70% ethanol and then resuspended in 40µl Tris EDTA buffer. The DNA was semi quantified by gel electrophoresis on a 1% agarose gel in 1xTAE buffer and visualised by under UV by ethidium bromide staining according to the procedure of Sambrook and kept at -200C for future use.14
The16S rRNA gene sequences were amplified with bacterial primer pair 8F forward 5’-AG (A/G) GTTTGATCCTGGCT-3’) and 1492R reverse, 5’-CGGCTACCTTGTTACGACTT-3’ (Sigma),15,16 using a PTC-100 thermal cycler mode (MJ research inc., USA). The PCR reaction mixture consisted of 40µl mixture containing 0.25µl of genescript Taq, 1.0µl (5-pmol) of 27F forward primer, 1.0µl (5-pmol) of 1492R reverse primer, 1µl of template DNA, 2.5 ul of dNTPs mix (2.5mM), 4.0 PCR 10x buffer (genescript) and 30.25µl of PCR water. The PCR cycling consisted of an initial 5 minute enzyme activation at 94oC followed by 35 cycles of a denaturation step at 94oC for 45 seconds, primer annealing step at 53oC for 60 seconds, chain elongation step at 72oC for 2 minutes and a final extension at 72oC for 5 minutes.16 The presence and concentration of amplified products was confirmed by ethidium bromide staining and visualization on 1% agarose gels in 1X TAE buffer (Figure 3). The PCR products were purified using the QIAquick PCR purification Kit protocol (Qiagen) and sent for sequencing at the International Livestock Research Insitute (ILRI), Nairobi, BecA-ILRI Hub Services>SegoliP sequencing facility.
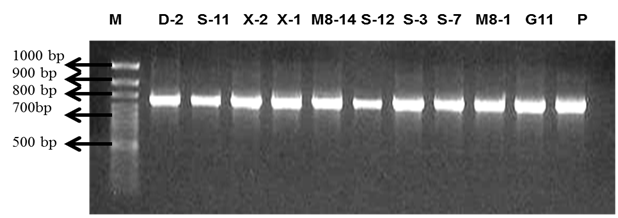
Figure 3 A photograph of PCR amplified 16S rDNA products of 10 isolates using Universal primers bac 8F and bac 1492-R, run in 1 % (W/V) agarose gel. M-Marker and P-positive control.
Sequencing and phylogenetic analysis of 16S rRNA sequences
Gene sequences were edited by eliminating all gaps (Complete deletion option), using the CHROMAS PRO software 1.5 version and compared to the sequences in the public databases using BLAST programme (www.ncbi.nih.gov/blast/blast.cgi) followed by alignment using CLUSTAL W 1.6 software.17 Phylogenetic trees were then constructed using the Neighbor-Joining method (Figure 4) (Figure 5). Evolutionary distances were calculated using the Jukes and Cantor (1969) method while Felsenstein, (1985) bootstrapping method was used to determine the accuracy of the tree constructed. These evolutionary analyses were conducted in Mega 5.
Sampling site conditions of Lake Magadi
The pH of the sampled sites ranged from 9-10.28, whereas temperature had a wide variation acroos the sampling sites that ranged from 39.40C to 51.90C. The conditions obtained within the sampling sites have been summarized in Table 6 below. These can be compared with water conditions of other soda lakes (Table 1).
Isolates obtained from Lake Magadi
The isolates obtained per media are summarized in Table 6 above. The highest number of isolates was recovered from the modified carbon based media (M1-M4), originally described by Duckworth et al.18 Very few isolates were detected from the mineral rich media described in (Table 6). Among carbon rich media, the glucose media had the highest number of isolates. However, it is the media rich in xylose that produced the highest diversity as it yielded representatives of all the three recovered genera.
Parameter |
Station 1 |
Station 2 |
Station 3 |
Media type |
Number of isolates obtained |
Lake Temperature |
44.50C |
39.40C |
51.90C |
Glucose |
9 |
Surrounding temp. |
38.00C |
38.00C |
38.00 C |
Starch |
6 |
pH |
10-Sep |
9.63 |
10.28 |
Cellulose |
6 |
M6 |
3 |
||||
Conductivity (EC) |
0.04 ms |
20.0 ms |
6.18ms |
Xylose |
5 |
TDS |
0.02mg/l |
0.2mg/l |
8.6mg/l |
DSC-97 |
3 |
Table 6 Physical parameters of the sampled sites during sampling
Morphological characteristics of the isolates
A total of 37 isolates were recovered during the study whose morphology was diverse: short rod (27 isolates), large rods (9 isolates) and coccus (1 isolate) (Table 7). Three isolates had red to pink coloured colonies, while the rest of the colonies were either cream (19) or white in colour (15). Thirty four (34) isolates were Gram positive, 2 Gram variable and only one was Gram negative.
Isolate |
Colony color |
Colony shape |
Gram reaction |
C1 |
Red-pink |
Large rods |
Gram positive |
C3 |
White |
Short rods |
Gram positive |
C4 |
White |
Large rods |
Gram positive |
C5 |
Cream |
Short rods |
Gram positive |
C6 |
Cream |
Short rods |
Gram positive |
C13 |
White |
Short rods |
Gram positive |
D2 |
White |
Large rods |
Gram positive |
D6 |
White |
Large rods |
Gram positive |
D9 |
White |
Short rods |
Gram positive |
M6-1 |
White |
Short rods |
Gram positive |
M6-7 |
Cream |
Short rods |
Gram positive |
M6-10 |
Cream |
Short rods |
Gram positive |
M8-1 |
White |
Large rods |
Gram positive |
M8-5 |
White |
Large rods |
Gram positive |
M8-12 |
White |
Large rods |
Gram positive |
M8-14 |
White |
Large rods |
Gram positive |
M8-15 |
White |
Short rods |
Gram positive |
S2 |
Cream |
Short rods |
Gram positive |
S3 |
Cream |
Large rods |
Gram positive |
S7 |
Cream |
Short rods |
Gram positive |
S10 |
Red-pink |
Short rods |
Gram positive |
S11 |
Cream |
Short rods |
Gram positive |
S12 |
Cream |
Short rods |
Gram positive |
X1 |
Cream |
Short rods |
Gram positive |
X2 |
Cream |
Short rods |
Gram positive |
X3 |
Cream |
Short rods |
Gram positive |
X5 |
Cream |
Coccus |
Gram negative |
X6 |
Cream |
Short rods |
Gram positive |
G2 |
White |
Short rods |
Gram variable |
G4 |
Cream |
Short rods |
Gram positive |
G8 |
Cream |
Short rods |
Gram positive |
G11 |
White |
Short rods |
Gram positive |
G12 |
Cream |
Short rods |
Gram positive |
G14 |
Cream |
Short rods |
Gram variable |
G15 |
White |
Short rods |
Gram positive |
G18 |
Red-pink |
Short rods |
Gram positive |
G20 |
Cream |
Short rods |
Gram positive |
Table 7 Morphological characteristics of the isolates
Enzyme production ability of the isolates
Fifteen isolates showed an ability to produce enzymes of various types. Of the enzyme producers, 47% were protease producers, 47% lipase producers and 73% produced amylase while others were multi enzyme producers (Table 5).
Physiological characterizaton of the isolates
Growth at different salinities: When cultured at different salinity [NaCl] levels, all the isolates, except two (M8-1 and X-3) grew within salinity range of 5%-30%). Thirty three (33) isolates grew optimumly at salinity of 10%, all grew optimumly at 12% and 15%, while only 11 grew optimumly at 20% (Table 8).
Isolate |
5% |
7% |
10% |
12% |
15% |
20% |
25% |
30% |
35% |
S2 |
+ |
++ |
+++ |
+++ |
+++ |
++ |
+ |
+ |
- |
S3 |
++ |
++ |
+++ |
+++ |
+++ |
++ |
++ |
+ |
- |
S7 |
+ |
++ |
+++ |
+++ |
+++ |
++ |
+ |
+ |
- |
S10 |
++ |
++ |
+++ |
+++ |
+++ |
++ |
++ |
+ |
- |
S11 |
+ |
++ |
+++ |
+++ |
+++ |
++ |
++ |
+ |
- |
S12 |
+ |
++ |
+++ |
+++ |
+++ |
++ |
++ |
+ |
- |
X1 |
+ |
++ |
+++ |
+++ |
+++ |
++ |
+ |
+ |
- |
X2 |
++ |
++ |
+++ |
+++ |
+++ |
+++ |
++ |
+ |
- |
X3 |
- |
++ |
+++ |
+++ |
+++ |
++ |
+ |
+ |
- |
X5 |
+ |
++ |
+++ |
+++ |
+++ |
++ |
++ |
+ |
- |
X6 |
+ |
++ |
+++ |
+++ |
+++ |
+++ |
+ |
+ |
- |
M8-1 |
- |
+ |
++ |
+++ |
+++ |
+++ |
++ |
+ |
- |
M8-5 |
+ |
++ |
+++ |
+++ |
+++ |
++ |
++ |
+ |
- |
M8-12 |
+ |
++ |
+++ |
+++ |
+++ |
++ |
++ |
+ |
- |
M8-14 |
+ |
++ |
+++ |
+++ |
+++ |
++ |
+ |
+ |
- |
M8-15 |
+ |
++ |
+++ |
+++ |
+++ |
+++ |
+ |
+ |
- |
G2 |
+ |
++ |
+++ |
+++ |
+++ |
++ |
++ |
+ |
- |
G4 |
+ |
++ |
+++ |
+++ |
+++ |
++ |
+ |
+ |
- |
G8 |
+ |
++ |
+++ |
+++ |
+++ |
++ |
++ |
+ |
- |
G11 |
+ |
++ |
++ |
+++ |
+++ |
+++ |
++ |
+ |
- |
G12 |
+ |
++ |
++ |
+++ |
+++ |
+++ |
++ |
+ |
- |
G14 |
+ |
++ |
++ |
+++ |
+++ |
+++ |
++ |
+ |
- |
G15 |
+ |
+++ |
+++ |
+++ |
+++ |
+++ |
++ |
++ |
- |
G18 |
+ |
++ |
+++ |
+++ |
+++ |
+++ |
+ |
+ |
- |
G20 |
+ |
++ |
+++ |
+++ |
+++ |
+++ |
++ |
++ |
- |
C1 |
+ |
+++ |
+++ |
+++ |
+++ |
++ |
+ |
+ |
- |
C3 |
+ |
++ |
+++ |
+++ |
+++ |
++ |
++ |
+ |
- |
C4 |
+ |
++ |
+++ |
+++ |
+++ |
+++ |
++ |
+ |
- |
C5 |
+ |
++ |
+++ |
+++ |
+++ |
+++ |
++ |
+ |
- |
C6 |
+ |
++ |
+++ |
+++ |
+++ |
++ |
++ |
+ |
- |
C13 |
+ |
++ |
+++ |
+++ |
+++ |
++ |
+ |
+ |
- |
D2 |
+ |
++ |
+++ |
+++ |
+++ |
++ |
++ |
+ |
- |
D6 |
++ |
++ |
+++ |
+++ |
+++ |
++ |
+ |
+ |
- |
D9 |
++ |
+++ |
+++ |
+++ |
+++ |
++ |
++ |
+ |
- |
M6-1 |
+ |
++ |
+++ |
+++ |
+++ |
++ |
+ |
+ |
- |
M6-7 |
++ |
+++ |
+++ |
+++ |
+++ |
++ |
++ |
+ |
- |
M6-10 |
+ |
++ |
+++ |
+++ |
+++ |
++ |
++ |
+ |
- |
Table 8 Tolerance to different salinity levels by the isolates
Key, +growth took ≥5 days; ++ growth took 3-4 days and +++ growth took ≤2 days.
Growth at different temperatures: The isolates exhibited growth at temperature ranges of 15- 45°C, with optimal growth evidenced at between 30oC and 35oC (Table 2).
Growth at different pH levels
Most of the isolates grew at pH ranges of between 6.0-11.0 (Table 4). The number of isolates that grew optimumly at various levels of pH were as follows: 4 at pH of 7.5, 27 at pH 8.0, 31 at pH 9.0 and 11 at pH of 10.0. The biochemical characteristics of the isolates are as indicated in Table 8 below. Generally Most of the isolates were catalase and oxidase positive.
Molecular characterisation of the isolates
A sample of the gel electrophoresis results obtained in the study is represented in Figure 3. Though partial sequencing was done for all the isolates obtained in the study, thirty six isolates were blasted (Table 9). Of the isolates blasted, only twenty one (21) were placed into phylogenetic trees (Figure 4) (Figure 5). Blasting and the resultant phylogenetic analysis clustered the obtained isolates with three main genera: genus Bacillus (Bacillus subtilis, Bacillus cereus and Bacillus licheniformis), Genus Clostridium (Clostridium sporogenes and Clostridium botulinum) and genus Halomonas (Halomonas campisalis). Bacillus relatives constituted 54% of the isolates followed by Clostridium at 38% and Halomonas at a mere 8%. The majority were Gram positives affiliated to genera of Bacillus and Clostridium) while the only Gram negative isolate belonged to the genus Halomonas.
Isolate |
Length |
Nearest relatives |
Accession number |
% similarity |
C4 |
664 bp |
Bacillus sp. W8B-43 |
HR238689 |
99% |
C1 |
862 bp |
B. subtilis strain Bs 20 |
HQ267754 |
78% |
C3 |
659 bp |
Bacillus sp. DB110 (2010) |
HM566824 |
98% |
C5 |
571 bp |
C.sporogenes strain McClung2004 |
NR029231 |
84% |
C6 |
873 bp |
Clostridium sp.SA-4 |
AY695835 |
95% |
D2 |
775 bp |
B. cereus strain PM-2 |
FJ598436 |
99% |
D6 |
672 bp |
B. cereus strain OCOR1DBT |
JF264468 |
99% |
D9 |
560 bp |
B. licheniformisstrain C18 |
HQ336641 |
99% |
G2 |
540 bp |
B. licheniformisstrain C18 |
HQ336641 |
99% |
S10 |
606 bp |
B. licheniformisstrain PRL1 |
EF062991 |
99% |
G4 |
581 bp |
Closridium sp. N56 |
AB600535 |
99% |
G8 |
581 bp |
Clostridium sp. N56 |
AB600535 |
99% |
G11 |
691 bp |
Bacillus sp. JM4 |
EF062991 |
99% |
G12 |
863 bp |
C. botulinum strain H04402 065 |
Dq196620 |
97% |
G15 |
596 bp |
B. licheniformisstrain GLU113 |
FN678352 |
100% |
G18 |
571 bp |
B. licheniformisstrain APS2MSU |
HQ637792 |
99% |
G20 |
764 bp |
C. sporogenes |
AB595130 |
85% |
G14 |
1021 bp |
C. sporogenes |
NR 029231 |
80% |
X1 |
731 bp |
Clostridium sp. SA-4 |
AY695835 |
99% |
X2 |
765 bp |
Clostridium sp. N56 |
AB600535 |
99% |
X3 |
786 bp |
C. botulinum strain 5564 |
HQ328066 |
88% |
X5 |
521 bp |
H. campisalis strain H10.1.1 |
GU228478 |
94% |
X6 |
847 bp |
B. licheniformis strain Crk21 |
GQ503331 |
97% |
S2 |
529 bp |
Clostridium sp. N56 |
AB600535 |
99% |
S3 |
714 bp |
Bacillus sp. OC-6 |
AY669167 |
99% |
S7 |
712 bp |
Clostridium sp. N56 |
AB600535 |
99% |
S12 |
716bp |
B. licheniformisstrain C18 |
HQ336641 |
99% |
S11 |
772 bp |
B. cereus strain SL3G |
HQ283476 |
99% |
M6-7 |
686 bp |
Clostridium sp. N56 |
AB600535 |
99% |
M8-1 |
692 bp |
B. cereus strain OCORIDBT |
FJ264468 |
99% |
M8-12 |
558 bp |
B. cereus EBT1 |
JF50734 |
99% |
M8-14 |
721 bp |
B. cereus strain OCORIDBT |
FJ264468 |
99% |
M8-15 |
674 bp |
Clostridium sp. N56 |
AB600535 |
99% |
M6-1 |
562 bp |
B. licheniformis strain APS2MSU |
HQ143636 |
99% |
C13 |
598 bp |
B. licheniformis strain BPRIST039 |
JF700489 |
99% |
M6-10 |
942 bp |
Bacillus licheniformis strain MEL09 |
FJ715927 |
98% |
Table 9 The blasted isolates, nearest neighbors in the data bank and their % relatedness
Growth at different physiological conditions
The ability of the isolated microbes to grow at 5% and above sodium chloride concentration confirms that they are true halophiles and not just halotolerants.1 All the isolates grew at [NaCl] of 20% and above and can therefore be considered extreme halophiles,1 even though they showed slow growth when sodium chloride concentrations rose above 15%, especially at [NaCl] of 25% and above where they took long to form colonies. Higher levels of salinity have been associated with dehydration, inhibition of proteolytic enzymes, and increased sensitivity to CO2. High salinity also removes O2 from the habitat. These could have contributed to observed low growth rates.
The observed optmal temperature ranges are within those bserved for microbes iosolated from other soda lakes. Halomonas magadii sp. nov., isolated from lake Magadi by Duckworth et al.,18 had an optimum temperature of 37oC, while the halomonad isolated by Bolyanskaya et al.19 from a USA alkaline lake had an optimum of 36-40ºC. Protease producing bacteria isolated by Abdelnasser et al.20 from Lake Wadi el-Natrun in Egypyt grew at temperature ranges of 25oC and 55ºC. In addition, the ability of these isolates to grow at temperatures lower than those recorded at sampling points (which ranged from 39.4oC and 51.9oC) is significant as it shows that these bacterial communities are adapted to survive over a wide range of temperatures.6
Most of the isolates grew at pH ranges of between 6.0-11.0 (Table 3). The abilty to grow at pH of 6 shows their abilty to tolerate a wide range of pH levels including mild acidity though they are of alkaline origin. Some of the documented pH ranges over which haloalkaliphiles and alkaliphiles grow include: 9.0 to 10,21 10-10.5,22 7.0 - 11.018 and 8 to 11.0.20 The observed pH values are therefore consistent with these earlier findings. However for the microbes that grew at pH 11.0, it took a longer period for the colonies to appear compared to growth at lower pH levels.
In general, the ability of the microbes isolated in this study to grow fast and optimally at the environmental and physicochemical parameters of high pH, elevated temperature and salinity unsuitable for the growth of other microorganisms makes Lake Magadi micro-organisms good models for research on adaptive mechanisms to extreme environmental parameters.7
Enzyme producing ability of Lake Magadi isolates
The ability of a number of the isolates, to produce various groups of intracellular enzymes such as gelatinase, catalase and citrate and extracellular enzymes, among them amylases, proteases and lipases (Table 3) (Table 5), even at the elevated conditions of pH and salinity of the lake water is indicative of their biotechnological potential. Again the production of the various groups of enzymes shows that the resident microbes are involved in the breakdown of the organic matter of the lake and its subsequent mineralization,23,24 and also in the nitrogen cycle of the lake, especially by isolates that showed an ability to reduce the nitrates. Indeed the presence of nitrogen reducers in any ecosystem is a necessary prerequisite for the completion of the nitrogen cycle. The microbes may therefore be major contributors in the transformation of organic carbon, sulfur and nitrogenous compounds and may also have an important role in the food webs and nutrient cycling of the lake ecology.
Molecular characterization and phylogenetic analysis of the isolates
The sequences that had more than 320 base pairs were blasted and consequently placed into phylogenetic trees. These are the sequences that could allow sensible alignment. The affiliation of the isolated microbes from Lake Magadi to the genera of Bacillus, Clostridium and Halomonas corresponds to databases of organisms discovered in the Rift Valley soda lake environment which contain records of members affiliated to these genera. The majority of the Gram positives were affiliated to the Bacillus and Clostridium genera while the only Gram negative isolate belonged to the genus Halomonas, a result that was in agreement with a similar study on Lake Magadi by.6 Duckworth et al.8 & Grant et al.23 also oberved that soda lakes contained more Gram positive bacteria of the wider Bacillus spectrum than Gram negative members of the family Halomonadaceae. The records show microbes recovered through both culture independent and dependent methods from littoral sediments of Lakes Magadi, Nakuru, Bogoria and Elmenteita.7,18,25 The dominance of relatives of genus Bacillus and Clostridium in other soda lake studies over the other groupshas also been reorted in previous studies. The abundance of Genus Bacillus is attributed to their endospore dormant stages that enable them to survive in the dry marginal areas around the lakes.23 An important finding of the study is the relationship of soda lake microbes to Bacillus subtilis, a common inhabitat of non-saline environments as it shows that many strains with similar 16S rDNA sequences are widespread in nature and can thrive under very different physiological conditions.26
The surprise finding however was the recovery of members affiliated to the genus Clostridium by the aerobic cultivation method used in this study. Genus Clostridium is thought to have members who are strict anaerobes.27 However Baumgarte6 isolated facultative species of Clostridium from Lake Magadi. McGraw-Hill also found out that Clostridia actually exhibit varying degrees of intolerance to oxygen. Some species of Clostridium are sensitive to O2 concentrations as low as 0.5%, most species are tolerant to O2 concentrations of 3-5% and few others grow minimally in air at atmospheric pressure.24 Those tolerant to O2 produce an antioxidant enzyme called superoxide dismutase (SOD).24 The sensitivity to O2 restricts Clostridia species to habitats that contain large amounts of organic matter as these provide optimal conditions for their growth and survival.27 This explains why the Clostridia isolates were only recovered from station 3 of the lake which is abundant in total dissolved solids (Table 6).
In the present study, all the enzyme secreting isolates were affiliated to the genus Bacillus. Previous Soda Lake studies that used direct plating on an agar media amended with substrates specific for enzymes of interest also isolated and characterized a number of enzyme secreting bacteria.2 Most enzyme activity was observed among members of the Gram positive genus Bacillus than in the Gram negative Halomonas species which produced enzymes such as amylases, proteases, lipases, DNases, pullulanases and xylanases. All the enzymes secreted by the isolates are thought to have remarkable potential in various industries, given that they are produced by inhabitants of an extremely saline and alkaline environment.
Whereas only two of the amplicons sequenced had 100% identity to sequences in the database, twenty nine isolates had a sequence similarity of 97% and above to their closest relatives and 7 had a sequence similarity of less than 97% to their closest relatives in the BLAST (Table 9). In fact some of the sequences such as X-3, G-12 and G-20 have very low identities (88% and below) to their respective relatives in the BLAST. Sequence identities of over 98% to a known organism represent the same species, an identity of between 88% and 98% is considered part of the same genus, while that of 88% and below like of the above isolates could represent new genera that have not been isolated and which may be specific to this habitat. This is the reasoning behind the use of such a wide range of media, modified with a variety of carbon sources and minerals whose intention was to widen the range of haloalkaliphilic organisms that would be cultured and promote the making of useful enzymes.8
Archaea are thought to be dominant in such hypersaline environments and the mineral rich media (Media 5 to 7) was included to specifically target members of Halobacteria. However none was obtained in the present study and though the initial morphological characterization of isolates S-10, G-18 and C-1 revealed the red and pink colony colours characteristic of halobacteria, phylogenetic analysis revealed that these were all related to genus Bacillus. The lack of archaeal diversity may be because only a few species of the class Halobacteria can grow above pH 1028 and the high sulfur content of soda lakes which might have favored the growth of chemosynthetic Bacteria and the fact that colonies of Archaea, may not have been picked from the agar plates during the purification. The halobacteria may also have lost the ability to form colonies on the particular agar plates used during repeated transfers in the purification procedures, or they may not have formed colonies because the media was unsuitable. The period of incubation might also have been too short to allow colonies of archea to form.
A lower diversity was obtained from this lake than anticipated. This can be attributed to the fact that two extreme conditions (salinity and alkalinity) were being investigated at the same time, which normally restricts the range and number of microorganism that can proliferate more than in a single growth condition.23 Such environments have a relatively restricted range of alkaliphilic inhabitants, usually Bacillus or related species.23 There is also a possibility of inadvertently not picking colonies of other bacterial members from agar plates during purification process. Our study both describes the existence of a bacterial diversity in Lake Magadi and indicates the industrial potential of some of the cultures especially the enzyme producing isolates. It also advances the understanding of the microbial ecology of Lake Magadi.
Media and diversity of microbes recovered
The highest number of isolates was recovered from the modified carbon based media (1-IV), originally described by Duckworth et al.,8 while a low number of isolates was detected in the mineral rich media. The carbon rich media also seemed to favour the growth of members affiliated to genera of Halomonas and Bacillus. Media rich in organic carbon is therefore suitable for the growth of fast growing microbes such as Bacillus and Halomonas. Though the glucose media had the highest number of isolates, it is the xylose carbon media that had the highest diversity, as it yielded representatives of all the three recovered genera. The gram positives (Bacillus and Clostridium) were more diverse and abundant than the Gram-negative (Halomonas). This concurs with a cultivation-dependent study on Lake Magadi done by Baumgarte6. Members of genera Bacillus, Clostridium and Halomonas are recorded in a database of organisms discovered in the Rift Valley soda lake environment such as the littoral sediments of Lakes Magadi, Nakuru, and Bogoria and Elmenteita, by both culture independent and dependent methods.7,18,25 Though thought to be strict anaerobes,27 Clostridia sp. isolated from Lake Magadi are most likely facultative anaerobes.6 The present study which employed an aerobic culture method, recovered members of the genus. Clostridia actually exhibit varying degrees of intolerance to oxygen, and some are sensitive to O2 concentrations as low as 0.5%, most species tolerate concentrations of 3-5% and few others grow minimally in air at atmospheric pressure,24 because they produce an antioxidant enzyme called superoxide dismutase (SOD).24 The sensitivity to O2 restricts Clostridia species to habitats that contain large amounts of organic matter,27–31 which provide optimal conditions for their growth and survival.
The study shows that Lake Magadi contains useful bacteria of different genera. The ability of the microbes to survive under extreme conditions of temperature and salinity exhibits their extreme haloalkalphilic nature. Further studies should be designed to capture a wider diversity of biotechnologically useful microbes form the lake.
None.
The author declare that there is no conflict of interest.

©2018 Nyakeri, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.