Journal of
eISSN: 2376-0060


Research Article Volume 2 Issue 5
1PPD Tuberculin Department, Razi Vaccine & Serum Research Institute, Karaj, Iran
2Antimicrobial Resistance Research Center, Mashhad University of Medical Sciences, Mashhad, Iran
3Arak University of Medical Sciences, Arak, Iran
4Aerobic Bacterial Research and Vaccine Production Department, Razi Vaccine & Serum Research Institute, Karaj, Iran
Correspondence: Nader Mosavari, PPD Tuberculin Department, Razi Vaccine & Serum Research Institute, Karaj, Iran; Razi Vaccine & Serum Research Institute, Karaj, Iran, Tel -4503055, Fax -4552357
Received: April 04, 2015 | Published: April 22, 2015
Citation: Soleimanpour S, Mosavari N, Asl DH. Zoonotic tuberculosis caused by Mycobacterium bovis, central province, Iran. J Lung Pulm Respir Res. 2015;2(5):93–96. DOI: 10.15406/jlprr.2015.02.00054
Background and objectives: Although Mycobacterium tuberculosis (M. tuberculosis) causes most cases of human tuberculosis (TB), Mycobacterium bovis (M. bovis) is an important cause of TB infections in certain populations. Identification of M. bovis as a member of M. tuberculosis complex could be valuable in term of public health control measures. In search for cases of zoonotic TB we have performed an 18-month molecular epidemiology study on all patients presenting pulmonary tuberculosis admitted to public hospitals in Markazi province, central Iran.
Materials and methods: Only one Mycobacterium bovis isolate was detected among 42M. tuberculosis isolates that were successfully collected during the research. Identified as M. bovis by phenotypic tests, PCR-RFLP for OxyR gene of this isolate coupled with findings of PGRS-RFLP and also DR-RFLP genotyping demonstrated an entirely different strain to those previously reported from animal hosts in Iran.
Results: Considering the very homogenous population of M. bovis in Iran, and the patient’s age, 75, we assume this isolate displays activation of an earlier exposure to M. bovis during the patient’s life.
Conclusion: Besides, our data suggest a low contribution of M. bovis in human TB in the region and possibly in the whole country.
Keywords: mycobacterium tuberculosis, mycobacterium bovis, zoonotic TB, PCR-RFLP, OxyR gene, RFLP
TB, tuberculosis; M. Bovis, mycobacterium bovis; M. tuberculosis, mycobacterium tuberculosis; MTBC, mycobacterium tuberculosis complex; HIV, human immunodeficiency virus
Tuberculosis still remains a global health problem. Most cases of disease are reported as Mycobacterium tuberculosis complex, while identification of Mycobacterium bovis as a member of this complex could be valuable in term of public health control measures. Although most cases of human tuberculosis are due to M. tuberculosis a proportion, called zoonotic TB, is caused by M. bovis as almost 3% of tuberculosis cases in the world are attributed to M. bovis.1 Human to human transmission of zoonotic TB is a rarely reported event2,3 and bovids (eg. Cattle) are the main hosts for M. bovis therefore, contact with infected animals or infectious animal products are blamed most in zoonotic TB cases.2,4 Prevalence of bovine tuberculosis is exacerbated by the presence of multiple additional risk factors such as high prevalence of human immunodeficiency virus (HIV) infections. However, HIV/AIDS facilitates transmission of any form of TB, some studies showed a significantly increased proportion of Mycobacterium bovis infections among HIV–co-infected TB patients compared with HIV-negative TB patients.5 According to the World Health Organization report, the prevalence of tuberculosis in Iran in 2013 was estimated 33 cases per 100000 people.6 On the other hand, Iran is among the seven Asian States with an existing bovine TB control scheme operating at national scale with a very homogenous of bovine M. bovis. This appears in concordance with few previously reported observations7 that cases of zoonotic TB in Iranian population is lower compared to its neighboring countries.8 Different approaches to differentiate M. bovis from M. tuberculosis complex are done but the PCR-RFLP using OxyR gene is highly sensitive and inexpensive method. The aim of this research is improving our epidemiological understand of M. bovis infection in human subjects in Iran. We performed a cross-sectional molecular epidemiology study on pulmonary tuberculosis patients admitted to a general hospital in central province of Iran to investigate zoonotic tuberculosis. During the study period forty two MTBC (Mycobacterium tuberculosis complex) isolates were identified by phenotypic tests, PCR-RFLP OxyR gene and genotypic methods including PCR-RFLP. Using IS6110-PCR, PCR-RFLP for OxyR gene plus RFLP (with PGRS and DR markers) genotyping strategies, the present research represents an epidemiological investigation into zoonotic TB in Central Province, Iran.
Study population: 41 Sputum specimens and one gastric lavage were collected during the period of March 2009 to September 2010 from all smear-positive TB patients (e.g. 38 Iranians and 4 Afghans) admitted to the public hospitals across the Markazi province, central Iran (Figure 1). In this study, five standard strains of mycobacterium including three strains of Mycobacterium tuberculosis (C, DT, PN), one strain of Mycobacterium bovis (AN5) and one strain of Mycobacterium bovis BCG (1173P2) were used for comparison.
Preferential growth of bacteria on LJ medium containing glycerol as compared with pyruvated LJ medium: The collected specimens were processed and cultured on glycerinated and also pyruvated traditional Lowenstein-Jensen (LJ) slopes according to the available standard protocols for preliminary identification of M. bovis isolates.9,10
Genomic experiments
Genomic DNA preparations: For 16SrRNA-PCR, IS6110-PCR, PGRS-RFLP and DR-RFLP tests the high quality genomic DNA was extracted as previously described by Van Soolingen.11
PCR-16SrRNA: The method of Huard was employed to amplify a 543bp-long fragment of the 16SrRNA using primer pair shown in Table 1.12
Target Gene |
Primer |
Oligonucleotide Sequence (5´-3´) |
Unit Size |
Anealing |
References |
16SrRNA |
F |
ACGGTGGGTACTAGGTGTGGGTTTC |
543 |
62 |
12 |
R |
TCTGCGATTACTAGCGACTCCGACTTCA |
||||
IS6110 |
F |
CCTGCGAGCGTAGGCGTCGG |
123 |
68 |
13 |
R |
CTCGTCCAGCGCCGC |
||||
Oxy R |
F |
GGTGATATATCACACCATA |
548 |
55 |
14 |
R |
CTATGCGATCAGGCGTACTTG |
Table 1 The primer sequences used
PCR-IS6110: The method of McHugh was employed to amplify a 245bp-long fragment of the IS6110 marker using INS1 and INS2 primers (Table 1).13
PCR-RFLP analysis of the OxyR gene: To differentiate between Mycobacterium bovis and M. tuberculosis within the MTB complex using the method proposed by Sreevastan, a 548bp genomic fragment of the OxyR gene was amplified by PCR using primers shown in Table 1. Then PCR product digested by AluI (Roche, Germany) and the fragments were separated by agarose gel electrophoresis.14 The following cyclic conditions were used for amplification of the oxyR gene: Denaturation at 94°C for 1min, 35 cycles of 21 s at 94°C, 21 s at 55°C, and 22 s at 72°C, and a final extension for 5 min at 72°C. DNA gel electrophoresis was carried out to test the amplification using 1.8% agarose gel under 90 V for 45 min. The PCR product (10μl) was digested with 4U of AluI. The reaction mix included 12μl water, 2.5μl enzyme buffer, and 10μl PCR product. A 100-bp marker was used as a ladder.
PGRS-RFLP and DR-RFLP: Internationally standardized protocol was employed to conduct these RFLP experiments.11,15,16 The achieved RFLP patterns were carefully observed by two experienced members of the research team and analyzed using GelPro® (MediaCybernetics, Millan, Italy).
Out of the 42 specimens (e.g. 42 patients) incorporated in the study; bacterial culture was successfully resulted in collection of all mycobacterial isolates. 41 isolates had better growth on glycerinated LJ compared with pyruvated LJ that is a sign of Mycobacterium tuberculosis. Only one isolate was grown in a pyruvated LJ stronger than glycerinated LJ that showed it is Mycobacterium bovis. In 16SrRNA-PCR, all isolates produced a 543bp-long fragment specific to mycobacterium genus and in IS6110-PCR all these isolates produced a 245bp-long typical fragment specific to members of M. tuberculosis complex (Figure 2). Given that, there are three AluI restriction sites in the 548-bp sequence of oxyR in M. tuberculosis complex organisms.14 In PCR-RFLP, about 41 isolates, digestion of the 548bp fragment of OxyR gene with AluI generated four DNA fragments of 236, 227, 55, and 30bp. However, the electrophoretic conditions showed only one band at about 230bp for DNA samples from M. tuberculosis complex organisms. This band represents both the 236-bp and the 227-bp oxyR gene, while 55bp and 30bp fragments were run out of the gel. The M. bovis isolates have four restriction sites for AluI in the amplified oxyR gene,14 and therefore, five DNA fragments (236, 148, 79, 55, and 30bp) were produced. However, the electrophoretic conditions used identified a three-band pattern of 236, 148, and 79bp for M. bovis isolates, while 55bp and 30bp fragments were run out of the gel (Figure 3). The PGRS-RFLP and DR-RFLP finding of the M. tuberculosis isolates have been presented and submitted for publication elsewhere. This was entirely different to those of M. tuberculosis isolates in the study setting. In RFLP, genetic patterns of M. bovis isolate were again entirely difference with M. tuberculosis isolates and didn’t have any similarity with previously reported genotypes from Iran (Figure 4).
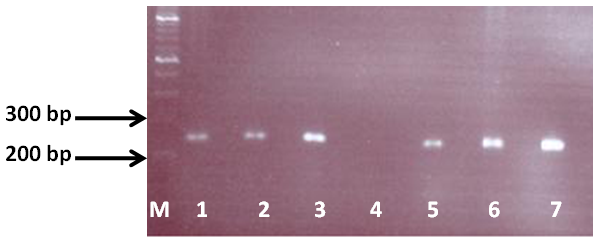
Figure 2 Electrophoretic fractionation of IS6110-PCR product in 1.5% agarose: M. tuberculosis complex isolates showed a band (245-bp). Lane M: DNA size standard, Lane 1, 2 and 3: M. tuberculosis standard strains (C, DT and PN), Lane 4: Negative control, Lane 5: M. bovis standard strain (AN5), Lane 6: M. bovis BCG (1173P2), Lane 7: human M. bovis isolate.

Figure 3 PCR-RFLP for OxyR gene after digested by the AluI: M. bovis isolate showed three bands (236, 148 and 79-bp) compared to reference strain, while 230-bp fragment was generated from M. tuberculosis. Lane 1: human M. bovis isolate, Lane 2: M. bovis standard strain (AN5), Lane 3: M. bovis BCG (1173P2), Lanes 4, 5 and 6: M. tuberculosis standard strains (C, DT and PN), Lane 7: Negative control, Lane 8: 100 bp DNA size standard.

Figure 4 RFLP patterns of M. bovis and M. tuberculosis isolates from central province of Iran using PvuII-digested DNA with the PGRS and DR probes. (A) DR probe: Lane 1: Standard size marker, Lane 2 and 3: M. tuberculosis isolates, Lane 4: human M. bovis isolate. (B) PGRS probe: Lane 1 and 2: M. tuberculosis isolates, Lane 3: human M. bovis isolate, Lane 4: Standard size marker.
Factors such as being neighbors with Tehran province, located on the main road access to the western areas of the country and a large number of traditional manufacturing and service units that are good purpose of non Iranian workforces including labor migrants and afghan asylum seekers leads to the fact that the Markazi province can accommodate a significant number of non-Iranian nationals. As a result, the transmission of infectious diseases like tuberculosis has changed epidemiological features of these diseases in this province.16 Until now there are only a few numbers of reports on zoonotic TB cases in Iran.17,18 Surprisingly, M. tuberculosis has not been yet found in cattle in this country7 although a rare case of infection is believed to be collected from buffalo recently in Urmiah, North-West of Iran.19
Finding of the present study are in agreement with previous findings as only one isolate in the study setting (N=42) was identified to be M. bovis. This finding suggests that M. bovis plays a minor role compared to M. tuberculosis in the etiology of pulmonary tuberculosis in Markazi province of Iran. A considerably large genetic diversity is seen in the population of M. tuberculosis in Iran confirmed mutually by almost all the recently employed DNA typing system20–23 while in contrast to this observation, a highly conserved population of M. bovis is reported from animals in Iran.7 The same scenario is represented by our findings as M. tuberculosis isolates proved highly polymorphic by either of RFLP methods (data not shown). We have looked into the database of bovine M. bovis genotypes already reported from Iran to see if the M. bovis genotype found in our study has been seen before in the country. Searching the RFLP databases of M. bovis available in Iran, we found no record of this isolate prior to the present study. When age of the patient hosting this specific isolate is taken into account, a 75years old female who has worked in a animal husbandry, possibility of infection at an earlier time due to exposure to a strain that for some unknown reasons is no longer frequent or available in cattle farms, appears to be explanatory as the most frequent bovine M. bovis strains have been exhaustively studied over the recent years in Iran. Therefore, we believe this is more likely to be a reactivation case of zoonotic tuberculosis rather than an epidemic transmission event as contact spoligotyping of this isolate.
To summarize, we assume zoonotic tuberculosis is currently not a major concern in this province of the country but further studies are suggested to extend the current information on biogeography of zoonotic TB in the whole nation.
The Razi Vaccine and Serum Research Institute (RVSRI) and the Medical Sciences University of Arak are thanked for funding and administrational supporting of the study. Doctors and laboratory staff at Karaj and Arak branches of RVSRI and also hospitals in Markazi province who participated in this piece of work are acknowledged. Mehdi Ahmadi, Shojaat Dashti and Samrand Reshadi are specifically thanked for their contribution in handling the clinical specimens. Saman Soleimanpour is a post graduate student at RVSRI.
The author declares no conflict of interest.

©2015 Soleimanpour, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.
 November is Lung Cancer Awareness Month, a vital opportunity for us to raise awareness about the dangers of lung cancer. Let’s unite to educate ourselves and others, and inspire proactive steps toward lung health. This year the motto of this event is “Stronger Together: United for Lung Cancer Awareness”. So for this occasion, the Journal of Lung, Pulmonary & Respiratory Research invites articles that emphasize the significance of lung protection. All the submissions received in the month of November will be offered with 40% discount on publication.
November is Lung Cancer Awareness Month, a vital opportunity for us to raise awareness about the dangers of lung cancer. Let’s unite to educate ourselves and others, and inspire proactive steps toward lung health. This year the motto of this event is “Stronger Together: United for Lung Cancer Awareness”. So for this occasion, the Journal of Lung, Pulmonary & Respiratory Research invites articles that emphasize the significance of lung protection. All the submissions received in the month of November will be offered with 40% discount on publication.