Journal of
eISSN: 2376-0060


Research Article Volume 8 Issue 2
1Department of Respiratory Medicine, 10th Zonal Tuberculosis and Chest Disease Center, Thailand
2Department of Pathology, Faculty of Medicine, Chiang Mai University, Thailand
Correspondence: Attapon Cheepsattayakorn, 10th Zonal Tuberculosis and Chest Disease Center, 143 Sridornchai Road Changklan Muang Chiang Mai 50100 Thailand
Received: December 24, 2020 | Published: April 1, 2021
Citation: Cheepsattayakorn A, Cheepsattayakorn R. Serological testing for COVID-19. J Lung Pulm Respir Res. 2021;8(2):35-39. DOI: 10.15406/jlprr.2021.08.00248
The objectives of this study are to identify the rapid, appropriate, screening, definite and novel methods of diagnosis of SARS-CoV-2 (COVID-19) infection, including SARS-CoV-2 (COVID-19) variants among various degree of COVID-19 severity for rapid prevention and control of SARS-CoV-2 (COVID-19) transmission. Methods of The Study: A comprehensive search was carried out in mainstream bibliographic databases or Medical Subject Headings, including ScienDirect, PubMed, Scopus, and ISI Web of Science. The search was applied to the articles that were published between 1971 and early March 2021. Results: With strict literature search and screening processes, it yielded 40 articles from 78 articles of initial literature database.
Characteristically, after infection, antibodies are detected in the blood of individuals, particularly individuals with few or mild symptoms. In patients with varying symptoms of COVID-19 and negative results of reverse-transcriptase-polymerase-chain reaction (RT-PCR) tests, the testing has a significantly clinical role when nasopharyngeal swabs are taken more than 5 days after symptom onset. The Royal College of Pathologists (RCPath) developed seven principles for production of a COVID-19 testing strategy. Testing being carried out for a purpose is one of these RCPath’s principles. Nevertheless, denial of requesting SARS-CoV-2 (COVID-19) antibody tests for reassurance should be cautioned. With a lower antibody levels, whether the protective immunity will be sustained is questionable. Several immune-based assays were developed against different SARS-CoV-2 (COVID-19) viral proteins as the followings: 1) Entire Spike (S) protein, IgG antibody from patient serum can cross-react with SARS-CoV and MERS-CoV, 2) S1 subunit of Spike (S) protein, IgA, IgG antibodies from patient serum can cross-react with SARS-CoV only, 3) Receptor-binding domain (RBD), IgG antibody from patient serum can cross-react with SARS-CoV only, and 4) Nucleocapsid (N), IgG antibody from patient serum can cross-react with SARS-CoV only. Long et al demonstrated in their study that IgG antibody and neutralizing antibody levels initiate decreasing within 2-3 months after infection in the majority of persons with recovery from SARS-CoV-2 (COVID-19) infection. An analytical study of the dynamics of neutralizing antibody titers demonstrated reduced neutralizing antibodies around 6-7 weeks after illness onset. In conclusion, the nucleic acid amplification tests may be poorly timed specimen collection, poor-quality specimen collection, long wait times for generating the results, and requirement of trained laboratory technicians. Serological data greatly supplement the laboratory results from the quantitative reverse-transcriptase-polymerase-chain reaction (qRT-PCR), the design of virus elimination programs (seroepidemiology), discovery of the monoclonal antibodies, and development of SARS-CoV-2 (COVID-19) vaccines.
Keywords: COVID-19, SARS-CoV-2, serological test, polymerase chain reaction, molecular diagnostics, variants, severe illnesses, complications
BAL: Bronchoalveolar Lavage, COVID-19: Coronavirus Disease 2019, CT: Computed Tomography, CRISPR: Clustered Regularly Interspaced Short Palindromic R epeat, IgA: Immunoglobulin A, IgG: Immunoglobulin G, IgM: Immunoglobulin M, MERS-CoV: Middle-East-Respiratory-Syndrome Coronavirus, N: Nucleocapsid, NPS: Nasopharyngeal Swab, qRT-PCR: quantitative Reverse-Transcriptase-Polymerase-Chain Reaction, RBD: Receptor-Binding Domain, RNA: Ribonucleic Acid, RT-LAMP: Reverse-Transcriptase Loop-mediated isothermal Amplification, RT-PCR: Reverse-Transcriptase-Polymerase-Chain Reaction, S: Spike protein, SARS-CoV-2: Severe Acute Respiratory Syndrome Coronavirus type 2, UK: United Kingdom, WHO: World Health Organization
The objectives of this study are to identify the rapid, appropriate, screening, definite and novel methods of diagnosis of SARS-CoV-2 (COVID-19) infection, including SARS-CoV-2 (COVID-19) variants among various degree of COVID-19 severity for rapid prevention and control of SARS-CoV-2 (COVID-19) transmission.
SARS-CoV-2 (COVID-19) is classified as a β-CoV of group 2B.1 The first notification of human COVID-19 occurred in Wuhan city, China and was reported by the World Health Organization (WHO) on December 31, 2019,1 whereas some experts hypothesized that the earliest case was detected on November 17, 2019.2. Subsequently, COVID-19 has rapidly spread through all continents and has reach the pandemic proportions contributing to declaration the Public Health Emergency of International Concern and global pandemic on January 30, 2020 and March 11, 2020, respectively.3 At the whole genome level, genetic analysis of SARS-CoV-2 demonstrated 92 % similarity to a bat coronavirus, BatCoV RaTG13.4 A previous study at whole genome level revealed that pangolin-CoV was identified to be 91.02 % similarity to SARS-CoV-2 (COVID-19).5 Pangolin and bat could be natural and intermediate hosts, respectively.4,5 Chronic respiratory diseases, hepatic diseases, cardiovascular diseases, malignancy, obesity, hypertension, septic shock, and diabetes are the risk of severe and critical COVID-19, particularly in the elderly6 that can contribute to acute respiratory distress syndrome (ARDS) requiring mechanical ventilation, renal injury, hepatic dysfunction, and multi-organ dysfunction or failure requiring intensive care support.7. A previous study demonstrated that infants can have a 7-10 % incidence of severe and critical COVID-19.8
Characteristically, after infection, antibodies are detected in the blood of individuals, particularly individuals with few or mild symptoms. In patients with varying symptoms of COVID-19 and negative results of reverse-transcriptase-polymerase-chain reaction (RT-PCR) tests, the testing has a significantly clinical role when nasopharyngeal swabs are taken more than 5 days after symptom onset.9, 10 The rate of RT-PCR detection of SARS-CoV-2 (COVID-19) in COVID-19 patients is 93 % in bronchoalveolar lavage fluid, 72 % in sputum, 63 % in nasopharyngeal swabs, 32 % in pharyngeal swabs, and 29 % in feces,11 whereas a previous small hospital cohort study demonstrated 15-30 % in blood and 14-38 % in rectal swabs.12 A recent meta-analysis of the sensitivity of the COVID-19 (SARS-CoV-2 viral RNA) diagnostic testing in saliva specimens in comparison to the sensitivity of the nasopharyngeal swab (NPS) tests demonstrated that the sensitivity for saliva tests was 91 % (CI=80-99 %), whereas the sensitivity of the NPS tests was 98 % (CI=89-100 %).13 Saliva could be an alternative valid strategy to serum for detecting antibodies against SARS-CoV-2 (COVID-19).
A comprehensive search was carried out in mainstream bibliographic databases or Medical Subject Headings, including ScienDirect, PubMed, Scopus, and ISI Web of Science. The search was applied to the articles that were published between 1971 and early March 2021. Our first involved performing searches of article abstract/keywords/title using strings of (“COVID-19” or “ SARS-CoV-2 ”, “ COVID-19 Variants ” or “ SARS-CoV-2 Variants ”, “ Methods of Obtaining Specimens For SARS-CoV-2 or COVID-19 Diagnosis ”, “ Screening Methods ”, “ Serological Diagnostics ”, “ Molecular Diagnostics ”, “ Diagnostic Methods ”, and “ SARS-Co-V-2- or COVID-19-Induced Severe Illnesses or Complications ”. After a first approach of search, published articles focusing on human COVID-19 were retained and the information on screening and diagnostic methods and COVID-19 severity was extracted for having a crude knowledge involving their themes. Another round of publication search was conducted for adding the missing published articles that were not identified by the first round.
All keywords combinations from one disease type and climatic variable to bind the population of cases under consideration. Search string for disease groups include. “ SARS-CoV-2 ” or “ COVID-19 ” or “ severity ” or “ screening methods ” or “ diagnostic methods ”. The initial literature databases were further manually screened with the following rules: 1) non-human COVID-19-related articles were excluded; 2)articles that did not report screening or diagnostic methods related to SARS-CoV-2 or COVID-19 were not considered, such as commentary articles, or editorial; 3)non-peer reviewed articles were not considered to be of a scholarly trustworthy validity; and 4) duplicated and non-English articles were removed. The articles were carefully selected to guarantee the literature quality, which is a trade-off for quantity.
With strict literature search and screening processes, it yielded 40 articles from 78 articles of initial literature database. Needed article information was extracted from each article by: 1) direct information including journal, title, authors, abstract, full text documents of candidate studies, publishing year; 2) place name of the study area; 3) study period; 4) research method used; 5) type of SARS-CoV-2 or COVID-19 screening or diagnostic variables studied; and 6) the conclusions made about the yields of screening or diagnostic methods on human COVID-19.
The availability and accuracy of antibody tests
Immunoglobulin M (IgM) rises soonest, whereas IgA and IgG persist. IgG alone. The maximum sensitivity for IgM alone, IgA alone, and IgG alone appear during the days 15-21 after the symptom onset that are 75.4 % (64.3-83.8), 98.7 % (39.0-100), and 88.2 % (83.5-91.8), respectively, whereas the specificity at all times for IgM alone and IgG alone are 98.7 % (97.4-99.3) and 99.1 % (98.3-99.6), respectively. The sensitivity and specificity of the antibody tests are critical due to false negative rates of RT-PCR that are between 2 % and 29 %.14
A previous study on immunological assessment of SARS-CoV-2 (COVID-19) infections in China conducted by Long et al. revealed that 81.1 % (30/37) and 62.2 % (23/37) of asymptomatic individuals tested positive for IgG and IgM, respectively and 83.8 % (31/37) and 78.4 % (29/37) of the symptomatic patients tested positive for IgG (around 3-4 weeks after COVID-19 exposure) and IgM, respectively. In acute phase that the viral ribonucleic acid (RNA) can be identified in a respiratory sample, IgG levels in symptomatic patients were significantly statistical higher than those in the asymptomatic individuals.15
Serological test interpretation
The pre-test probability of infection has much influence on the interpretation of the serological test results not only influenced by the accuracy of the test itself. When screening suggestive symptomatic individuals, the pre-test probability will be much higher, compared to asymptomatic persons.16
Serological testing pitfalls
COVID-19 screening is essentially amounted by non-specific indication and population-based policies on testing. In consequences of testing with uncareful consideration, this risks the potential harm. In more affluent populations, the rates of testing will be higher.17 that limits the estimates of seroprevalence. The Royal College of Pathologists (RCPath) developed seven principles for production of a COVID-19 testing strategy. Testing being carried out for a purpose is one of these RCPath’s principles.18. Denial of requesting SARS-CoV-2 (COVID-19) antibody tests for reassurance should be cautioned.19, 20
Immunity and antibody tests
In eliminating COVID-19, a combination of B and T cell immunity is likely to involve for production of protective-immunity memory.16. Nevertheless, currently, several longitudinal studies demonstrated waning of antibody levels.21. With a lower antibody levels, whether the protective immunity will be sustained is questionable.16 A recent study revealed that produced antibodies can provide long-term immunity, whereas non-neutralizing antibodies can be generated. Antibody enhancement, a phenomenon that can facilitate a more severe-secondary infection. This phenomenon is not to date with SARS-CoV-2 (COVID-19),but it has been demonstrated in other coronaviruses.22
Immune-based Assays developed against different SARS-CoV-2 (COVID-19) viral proteins
Several immune-based assays were developed against different SARS-CoV-2 (COVID-19) viral proteins as the followings: 1) Entire Spike (S) protein, IgG antibody from patient serum can cross-react with SARS-CoV and MERS-CoV, 2) S1 subunit of Spike (S) protein, IgA, IgG antibodies from patient serum can cross-react with SARS-CoV only, 3) Receptor-binding domain (RBD), IgG antibody from patient serum can cross-react with SARS-CoV only, and 4) Nucleocapsid (N), IgG antibody from patient serum can cross-react with SARS-CoV only.23.
Novel diagnostic methods for detecting SARS-CoV-2 (COVID-19) variants
Recently, there were variations of SARS-CoV-2 (COVID-19) developed and transmitted in the United Kingdom (UK) (201/501Y.V1/B1.1.7), South Africa (20H/501Y.V2/B.1.351), and Brazil (P.1/201/501Y.V3/B.1.1.248). The SARS-CoV-2 (COVID-19) variants in the UK can be detected by current molecular methods, such as qRT-PCR,24 but there are no data on impact on molecular assay performance for detection SARS-CoV-2 (COVID-19) variants in South Africa and Brazil. SARS-CoV-2 (COVID-19) variants may impact molecular assays that target S gene sequence of SARS-CoV-2 (COVID-19).25. There are no data on impact on serological antibody tests for detecting SARS-CoV-2 (C)OVID-19) variants in the UK, South Africa, and Brazil, but there is potential for the performance of a assay detecting antibodies to viral spike protein or nucleocapsid to be affected.25 Considering the performance of antigen-based tests (including rapid lateral flow devices), five SARS-CoV-2(COVID-19) rapid antigen tests are all able to detect the SARS-CoV-2ID-19) variants in the UK,24 but no evaluation studies are available for detecting SARS-CoV-2COVID-19) variants in South Africa and Brazil.25 Recently, additional novel assays based on the isothermal amplification of viral nucleic acids, in combination with clustered regularly interspaced short palindromic repeat (CRISPR)-based detection methods have been developed. These methods do not require thermal cycling and are more rapid than RT-PCR and are considered as point-of-care tests for SARS-CoV-2 (COVID-19) detection.26-28
IgG antibody responses sustained for at least 34 months after outbreak in persons with laboratory-confirmed MERS-CoV infection,29 whereas IgG levels in SARS-CoV-infected individuals were sustained for more than two years.30,31 Neutralizing antibodies that associate with the numbers of virus-specific T cells have been detected in most COVID-19 convalescent patients.32-35 Long et al. demonstrated in their study that IgG antibody and neutralizing antibody levels initiate decreasing within 2-3 months after infection in the majority of persons with recovery from SARS-CoV-2 (COVID-19) infection.15 An analytical study of the dynamics of neutralizing antibody titers demonstrated reduced neutralizing antibodies around 6-7 weeks after illness onset.36 Isothermal amplification is a useful alternative to thermal-cycling-based nucleic acid amplification.37, whereas RdRp/Hel assays are highly sensitive methods for detection of SARS-CoV-2 (COVID-19).38 Other novel diagnostic technologies for detecting SARS-CoV-2 (COVID-19) are nanotechnology-based-reverse-transcriptase loop-mediated isothermal amplification (RT-LAMP).39 and multiplex SARS-CoV-2 antibody immunoassays.40 Variation of the levels of SARS-CoV-2 (COVID-19) RNA and antigen, IgM and IgG antibodies after infection and clinical interpretation of the molecular and serological tests are demonstrated in Figure 1 and Table 1, respectively.41 The conventional chest radiography provides the sensitivity approximately 60 % for initial detection of pulmonary diseases in 1,014 COVID-19 patients,42 whereas the sensitivity of the chest computed tomography (CT) for COVID-19-related pulmonary diseases was 97 % among positive RT-PCR-COVID-19 patients and 75 % of COVID-19 patients with negative RT-PCR results revealed positive chest CT scans (308 of 413 patients).43 Thus, chest CT scans,44 and chest radiography.45 could be adjunctive tests, in combination with repeated RT-PCR assays and serological tests.
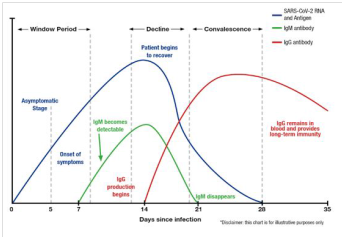
Figure 1 Demonstrating the levels of SARS-CoV-2 (COVID-19) RNA and antigen, IgM and IgG antibodies after infection.41
|
Test Results |
Clinical Interpretation |
||
|
RT-PCR |
IgM |
IgG |
|
|
positive |
negative |
negative |
May be in the window period of infection |
|
positive |
positive |
negative |
Early-stage of infection |
|
positive |
positive |
positive |
Active phase of infection |
|
positive |
negative |
positive |
Late phase or recurrent stage of infection |
|
negative |
positive |
negative |
Early stage of infection or false positive for RT-PCR |
|
negative |
negative |
positive |
Recovered |
|
negative |
positive |
positive |
Recovery phase or false positive for RT-PCR |
Table 1 Demonstrating clinical interpretation of molecular and serological tests.41
Serological data greatly supplement the laboratory results from the quantitative reverse-transcriptase-polymerase-chain reaction (qRT-PCR), the design of virus elimination programs (seroepidemiology), discovery of the monoclonal antibodies, and development of SARS-CoV-2 (COVID-19) vaccines. The nucleic acid amplification tests may be poorly timed specimen collection, poor-quality specimen collection, long wait times for generating the results, and requirement of trained laboratory technicians.
Dr. Attapon Cheepsattayakorn conducted the study framework and wrote the manuscript. Associate Professor Dr. Ruangrong Cheepsattayakorn contributed to scientific content and assistance in manuscript writing. Both authors read and approved the final version of the manuscript.
None.
The authors declare that they have no actual or potential competing financial interests.

©2021 Cheepsattayakorn, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.
 World Tuberculosis Day (March 24) provides the opportunity to raise awareness about TB-related
problems and solutions and to support worldwide TB-control efforts. While great strides have been made to control and cure TB, people
still get sick and die from this disease in our country. On this event, we request researchers to spread more information and awareness
on this by their article submissions towards our JLPRR. For this we are rendering 25% partial waiver for articles submitted on or before
March 24th.
World Tuberculosis Day (March 24) provides the opportunity to raise awareness about TB-related
problems and solutions and to support worldwide TB-control efforts. While great strides have been made to control and cure TB, people
still get sick and die from this disease in our country. On this event, we request researchers to spread more information and awareness
on this by their article submissions towards our JLPRR. For this we are rendering 25% partial waiver for articles submitted on or before
March 24th.