Journal of
eISSN: 2376-0060


Research Article Volume 9 Issue 3
Cancer institute and hospital, Tianjin medical university, China
Correspondence: Ruiping Zhang, Tianjin Medical University, Cancer Institute and Hospital, China
Received: November 11, 2022 | Published: November 25, 2022
Citation: Zhang R. Nomogram of baseline CT-radiomics from small-cell lung cancer patients: evaluation of added prognostic value for overall survival and probability of distant metastasis. J Lung Pulm Respir Res. 2022;9(3):79-85. DOI: 10.15406/jlprr.2022.09.00285
Objectives: To evaluate the added prognostic value of baseline CT-radiomics using nomogram for overall survival and probability of distant metastasis in small-cell lung cancer patients.
Materials and methods: This retrospective study consisted of 122 patients with stage IIA-IIIB small-cell lung cancer,which 97 patients for training dataset and 25 for validation dataset. The function defined as rad_score was constructed by the linear combination of selected radiomics features from baseline CT images weighted by their respective logistic coefficients and intercept in the LASSO-Cox model. The nomogram was developed based on the above function for overall survival (OS) and calibrated by the Harrell’s concordance index (C-index). The performance of the classifiers for DM was evaluated by receiver operating characteristics (ROC) curves with the indictor of area under curves (AUC). Furthermore, survival curve depicted by Kaplan-Meier method was compared with Log-rank test between low- and high-risk group.
Results: The nomogram performance of radiomics features and risk clinical factors (c-index of 0.64) don’t take advantage over the one of risk clinical factors-based alone (c-index of 0.596). The probability prediction of combination of the radiomics and clinical risk factor, radiomics alone, and clinical factors alone was shown, namely AUC of 0.673, 0.640 and 0.650, respectively. No significant different was found between ROCs (p-value > 0.4, Delong test). Moreover, we compared the Kaplan-Meier curves between low- and high-risk group, and showed p<0.001 with Log-rank test.
Conclusion: In the study, we can not confirm the hypothesis that baseline CT-radiomics contribute to predict the OS and probability of DM significantly. Moreover, the nomogram model based on combination of radiomics and clinical parameters has disadvantage over clinical parameters alone, probably affected by heterogeneity of datasets or SCLC need more valuable information for prediction outcomes.
Keywords: small cell lung cancer, distant metastasis, overall survival, radiomics
Baseline computed tomography (CT) imaging of before radiotherapy is routinely used for tumor segment and treatment plan designing. Although the CT images provide valuable information to guide treatment, the more quantitative information should be further extracted for the development of precision medicine.1,2 Radomics are able to quantify the information differing from medical images by using a huge number features that can be combined with clinical factors of tumor.1,3,4 The prognostic and predictive performance have been studied widely based on the baseline CT images using radiomics in non-small cell lung cancer (NSCLC).5–7 Besides that, potential added value of CT-radiomics during the radiotherapy has been explored for outcome prediction in both lung cancer and head-neck cancer.8,9 However, no related reports were published for outcome prediction for small cell lung cancer (SCLC).
According to the above concerns, without extra patient burden, we hypothesize that the combination both the baseline CT-radiomics and clinical parameters might provide comprehensive visual information to predict outcome for SCLC via nomogram model. A previous study showed that the convenient nomogram, which as a model for a scoring system, can be used to evaluate a model in which important prognostic factors can be combined for risk prediction for an individual patient.10,11 The scoring system can provide the most accurate predictions for specified end points and fulfils personal requirements which can produce individual survival estimations. Even the current estimated approach demand novel prognosticators that allow for further classifying the different risk groups. Nevertheless, the prognostic value of combination has not been explored, the patients with high risk might receive certain adjuvant therapy, not only improving the therapeutic effect, but also avoiding the over-treatment of low-risk patients and more refined therapeutic strategies. So in this study, the first goal is to predict the probability of DM based on a combination of risk clinical factor and optimal radiomics features, and the second goal is to build a scoring nomogram that incorporated both of the radiomics and risk clinical factors in order to estimate individual OS efficiently and conveniently.
Patient characteristics
This retrospective study was approved by the Tianjin Cancer Hospital Medical Ethics Committee and informed consent was waived. Two hundred thirty patients with pathology-confirmed small-cell lung cancer (SCLC) or composite SCLC12 were enrolled from 2011 to 2016 (overall stage II-III). The pathological diagnoses were reviewed again and confirmed independently by two pathologists, each of whom has at least fifteen years of experience. Patients treated with surgery or radiation therapy prior CT simulation date were excluded. Patients’ clinical characteristics were derived from medical records (gender, age, smoking status, clinical stage, pathology) according to our uniform guidelines.
Clinical follow-up
According to our routine clinical follow-up protocol, chest contrast-enhanced CT scans were required for each patient every three months after the first two-year treatment and every six months in the following five years. DM was defined as progress of tumor into other organs which were assessed from the CT image by radiologist and clinician, and the time to DM were defined as the date of the last scan for patients. OS time was defined as the time between the date of diagnosis and the date of death or the last follow-up date.
CT image acquisition and tumor segmentation
All of patients underwent contrast-enhanced chest CT image acquisition with free breathing (Brilliance CT big bore, Philips Healthcare, Best, Tshe Netherlands) with technique parameters set according to standard clinical chest scanning protocols. The slice thickness and pixel size of imaging was 3mm and 1.25 mm by 1.25 mm, respectively. The primary tumor was contoured by an experienced radiation oncologist on a treatment planning workstation (Pinnacle, Philips Radiation Oncology Systems, Fitchburg, WI). Two expert radiologists reviewed all slices of each patient and confirmed the primary tumor contours together.
Radiomics feature extraction
Image and primary tumor files of patients in Digital Imaging and Communication in Medicine (DICOM) format were retrieved and imported into a workstation platform (Radcloud, version 2.1.2, http://radcloud.cn/, Huiying Medical Technology Co., Ltd, Beijing, China) before extraction of radiomics features. Radiomics features of three groups were extracted. The three groups were as following: 1) first-order features, 2) shape features, 3) texture features. The further detailed information about radiomics features is shown in Appendix A.
Feature standardization
Before feature selection, each radiomics feature along with continuously variable clinical information was standardized independently by subtracting the mean values and scaling values to unit variance. This was done to avoid some features overwhelming others, while categorical features from clinical information were encoded as a numeric array using the one-hot encoding scheme.
Radiomics feature selection and rad_score construction
The least absolute shrinkage and selection operator (LASSO) method,13,14 which proved to be effective in regression of high-dimensional data,15–17 was used to select the most valuable radiomics features in our study. For the binary classification task of DM prediction, the original LASSO method was adequately competent to perform feature selection.
For the regression task of OS nomogram development, the LASSO method should be combined with a Cox proportional hazards model,18,19 to produce a LASSO-Cox regression model.
Due to the limited dataset size, the strategy of implementing LASSO/LASSO-Cox method consisted of two parts. First, 10-fold cross-validation of the whole dataset was used to select the optimal regularization constant α via the minimum of average mean square error (Appendix B). Then, the features with non-zero coefficients in the LASSO/LASSO-Cox models were selected based on the optimal value of α. Moreover, for the task of OS nomogram development, the rad_score function was calculated by the linear combination of selected radiomics features weighted by their respective coefficients and intercept in the LASSO-Cox model for each patient.
Clinical information selection
Due to the small number of factors in the clinical information set, the optimal factor subset was found by a complete search20 according to the minimum of mean Akaike’s information criterion (AIC)21 in the ten-fold cross-validation of the whole data set. In this process, all of the models were built by a logistic regression method.
The feature standardization and one-hot encoding and feature selection of radiomics and clinical information for the task of DM prediction was done using the Python 3.6 language (https://www.python.org/) in an Anaconda3 platform (https://www.anaconda.com/) with the scikit-learn software package (https://scikit-learn.org/), while the LASSO-Cox method for the task of OS nomogram development was performed with the R 3.4 language (http://www.R-project.org/) in a RStudio platform (https://www.rstudio.com/) and glmnet package (http://cran.r-project.org/web/packages/glmnet/).
Statistical analysis
Classifiers for DM probability prediction
The classifiers for DM prediction of SCLC in our study were developed using the method of logistic regression based on the selected radiomics features, clinical factors, and the combination of the two, with the strategy of five-fold cross-validation in the whole dataset. The predictive performance of those classifiers was evaluated by receiver operating characteristic (ROC) curves with the statistical indices of areas under the ROC curves (AUC), accuracy, specificity and sensitivity, and compared by the Delong test.22
Individualized nomogram for OS
The individualized nomograms for OS were built by the method of Cox proportional hazards regression based on the selected clinical factors and the combining both rad_score and the clinical factors, respectively, which were assessed by the Harrell’s concordance index (C-index)23 and calibration curves. Furthermore, the cut-off values of rad-score were determined by the median, separating the whole dataset into low- and high-risk groups. Survival curves for the whole dataset and the two groups dataset were depicted by the Kaplan–Meier (KM) method, and compared by use of the Log-rank test.22
The statistical analysis including model development and graphing for the task of DM probability prediction was also done using Python 3.6 in an Anaconda3 platform with the packages of scikit-learn and matplotlib (https://matplotlib.org/), while the analysis for the task of OS nomogram development were performed using R 3.4 in a RStudio platform with the packages of survminer (https://cran.r-project.org/package=survminer) for the KM method, rms (https://cran.r-project.org/package=rms) and regplot (https://cran.r-project.org/package=regplot) for nomogram building and C-index calculation. Of particular note is the plotting of calibration curves in Anaconda3 using Python and matplotlib for better customized graphing. P-values below 5% were considered statistically significant.
Study population
One hundred twenty-two SCLC patients treated with radiation therapy were included for our study. Of these eligible patients, 84 were males and 38 were females. Their median age was 58 ± 8.8 years. The median follow-up time was 19 months (range: 5.0-40.2 months). The median OS was 14 months. The median time to DM was 12.2 months with the 41.8%patients for which DM occurred, versus 58.2% who did not have DM. All of the patient characteristics and clinical outcomes are shown in Table 1.
Characteristics |
Total selected patients(N=122) |
Gender |
|
Male |
84 |
Female |
38 |
Age (y) |
|
≤60 |
78 |
61-69 |
35 |
≥70 |
9 |
Mean ± SD (y) |
58 ± 8.8 |
Clinical Stage n (%) |
|
IIA |
4 (3.3) |
IIIA |
61 (50) |
IIB |
7 (5.7) |
IIIB |
50 (41) |
Pathology n (%) |
|
Small Cell Lung Cancer |
112 (91.8) |
Composited Small Cell Lung Cancer |
10 (8.2) |
Smoking Status n (%) |
|
Yes |
82 (67.2) |
No |
40 (32.8) |
Follow-up close out date |
Mar-18 |
Follow-up for the entire study (months) |
|
Median (range) |
19 (5.0-40.2) |
OS (y) |
|
Median (range) |
14.82 (3.68-38.28) |
Mean ±SD |
16.03 ± 7.76 |
Distant metastasis n (%) |
51 (41.8) |
Table 1 Baseline characteristics of patient with small cell lung cancer (SCLC)Note: OS, overall survival; SD, standard deviation
Feature selection and rad_score construction
Clinical factors selected composed of the top three performing factors from all of the clinical factors. Three valuable prognostic clinical factors were selected for DM and OS. For DM, they are age, clinical stage, smoking status, whereas for OS, they are clinical stage, pathology, and smoking status.
The number of radiomics features was reduced to three features for DM and four for OS regarding prognostic stability and minimizing redundancy. Two of these features were texture-based and one was shape-based for DM, and three were texture-based and one was statistics-based (first-order) for OS.
The two rad_score functions can be constructed and computed as following:
For DM
Rad_score = Wavelet.LHL_GLSZM_LargeAreaLowGrayLevelEmphsis X
0.0261779 + Original_Shape_Maximum2DDiameterRow X
0.02370295 + Wavelet.LLH_GLSZM_ZoneEntropy X 0.000777
For OS
Rad_score = Wavelet.HLL_GLCM_IDMN X 0.09926913 +
Wavelet.HLL_First Order_InterquartileRange X 0.08657353 +
Square_GLRLM_RunEntropy X 0.04815856 +
Wavelet.HLL_GLCM_IDN X 0.00382770
A more detailed description of the selected features is provided in Table 2 and Appendix A.
|
Selected clinical factors |
Selected radiomics features |
Distant Metastasis |
Age |
Wavelet.LHL_GLSZM_LargeAreaLowGrayLevelEmphasis |
Clinical Stage |
Original_Shape_Maximum2DDiameterRow |
|
Smoking |
Wavelet.LLH_GLSZM_ZoneEntropy |
|
Overall Survival |
Clinical Stage |
Square_GLRLM_RunEntropy |
Pathology |
Wavelet.HLL_First Order_InterquartileRange |
|
Smoking |
Wavelet.HLL_GLCM_IDMN |
|
|
|
Wavelet.HLL_GLCM_IDN |
Table 2 A Potential parameters of clinical factors and radiomics features for predicting the probability of distant metastasis and overall survivalNote: GLCM, gray level co-occurrence matrix; GLSZM, gray level size zone matrix; GLRLM, gray level run length matrix; IDMN, inverse difference moment normalized; IDN, inverse difference normalized
Prediction for probability of DM
The prognostic performance of the combination of the radiomics and clinical risk factor, radiomics alone, and clinical factors alone was 0.673 (AUC; 95%CI, 0.583, 0.755), 0.640 (AUC; 95%CI, 0.548, 0.725), and 0.650 (AUC; 95%CI, 0.558, 0.734), respectively. No significant different was found between ROCs (p-value > 0.4, Delong test)22. The ROC curves are shown in Figure 3, and the assessment (accuracy, specificity and sensitivity) is listed in Table 3.
Predictive model |
AUC (95%CI) |
Accuracy |
Specificity |
Sensitivity |
radiomics-based |
0.640(0.54-0.74) |
0.639 |
0.746 |
0.49 |
clinical risk factors-based |
0.650(0.574-0.719) |
0.648 |
0.648 |
0.647 |
Combination both of radiomics and clinical risk factors |
0.673(0.533-0.679) |
0.607 |
0.606 |
0.608 |
Table 3 Predictive performance of radiomics-based model, clinical risk factors-based, and combination both of radiomics and clinical risk factors for distant metastasis (DM)Note: AUC, area under the curve; 95% CI, 95% confidence internal
Assessment of Nomogram for OS
Nomograms integrating clinical risk factors and rad_score, and of clinical risk factors alone were developed in Figure1a and Figure1b, respectively. The two nomograms assessed the probability of survival for 1 year, 15 months, and 2 years after radiotherapy. The combination model of rad_score and clinical risk factors was developed and shown in Figure 1a. Interestingly, rad_score has over-performed the significance on clinical risk factors, and clinical risk factors do not show any statistically significance with p-value > 0.05. However, clinical risk factors-based nomogram (Figure 1b), smoking status and clinical stage have outstanding performance (p-value<0.01). Incorporating the rad_score into the radiomics-based nomogram resulted in slightly better performance (C-index: 0.641; 95%CI: 0.61, 0.672) than which the clinical risk factors-based nomogram (C-index: 0.596; 95%CI: 0.593, 0.599). There is no significant difference between two calibration curves which is shown in Figure1c & 1d.
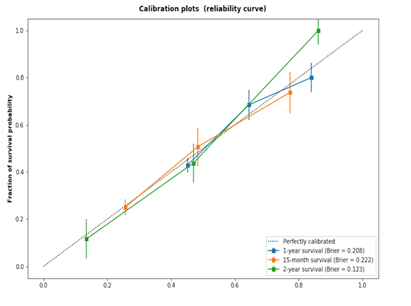
Figure 1d (C-index: 0.596) depict the consistency in terms of the agreement between the estimated survival probability and the observed survival probability.Source: Radiomics-based nomogram and clinical risk factors-based nomogram that estimate individual overall survival probability in patients with SCLC, along with the assessment of the model calibration. Radiomics-based nomogram (a) and clinical risk factors-based nomogram (a). For radiomics-based nomogram, both of patients’ rad_score and clinical risk factors are on the vertical axis. For clinical risk factors-based nomogram, clinical risk factors alone are on the vertical axis. Points are assigned for each risk factor by drawing a line upward from the corresponding values to the ‘points’ line. The total sum of points for four risk factors (a) or three risk factors (b) is plotted on the ‘total points’ line. A line is drawn down to the corresponding predictions of 1-year, 15-month and 2-year survival probability. Calibration plot for radiomics-based nomogram (c) (C-index: 0.641) and based for only risk clinical factors (d) (C-index: 0.596) depict the consistency in terms of the agreement between the estimated survival probability and the observed survival probability. The dotted line represents a perfect estimation by an ideal model, in which the estimated outcome fully meets the observed outcome. The solid line represents performance of the nomogram; the closer the line to the dotted line means more better estimation.
The association of the radiomics with survival was assessed by using Kaplan-Meier curve. All patients were stratified into as high- and low-risk groups by rad_score median. The KM curve of the whole dataset was also provided (Figure 2a), and median OS was fourteen months. From Figure 2b, the KM curve of OS was found to be clearly separated. The median OS of seventeen and twelve months for the low- and high-risk groups (n=122, Log-rank test, p-value < 0.0001), respectively.
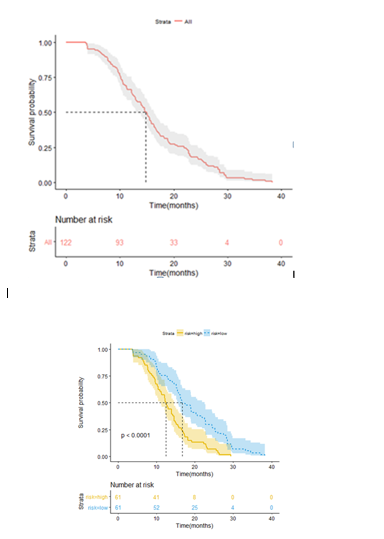
Figure 2 Illustration of Kaplan-Meier survival curves. KM plot of the whole patient dataset (a), and the median of OS is fourteen months. All the SCLC patient dataset were stratified into high- and low-risk groups using the cut-off of rad_score median (b). The yellow solid line is for KM plot of high-risk group, blue dotted line for KM plot of low-risk group. The median of OS is seventeen months for low-risk group and twelve months for high-risk group. A significant survival difference appears to be five months between the two groups (n=122, Log-rank test, p-value < 0.0001).
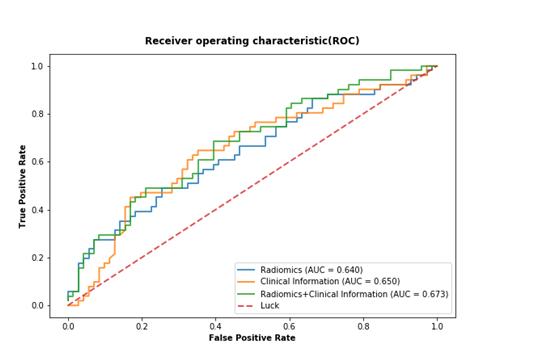
Figure 3 Receiver operating characteristic (ROC) curves for only radiomics-based model (blue solid line) (AUC=0.640, 95%CI, 0.548-0.725), the only clinical risk factors-based model (orange solid line) (AUC= 0.650, 95%CI, 0.558-0.734) and combined rad_score and clinical risk factors model (green solid line) (AUC= 0.673, 95%CI, 0.583-0.755) for prediction of DM in SCLC patients. The area under the curve (AUC) are shown in the lower right corner of the figure. No significant different is found between ROCs (all of p-values > 0.4, Delong test).
The goal of this study was to investigate the complementary prognostic value of baseline CT-radiomics features for overall survival and probability of distant metastasis in SCLC patients. We hypothesized that baseline radiomics features contain additional information in describing tumor characteristics. To this extent, significant radiomics features were explored, and nomogram models were developed using the significant features and risk clinical factors, calibrated, and assessed by c-index. Model performance of only significant features was compared to that of a model based on risk clinical factors. There classifier performance in probability of distant metastasis including lonely clinical risk factors and lonely radiomics, as well as both sets of factors with ROC curves using the AUC as a metric.
Probability prediction of distant metastasis
The patients with potential distant metastasis should be noticed to clinicians to pay additional medical care. In this study, we hypotheses that Radiomics features might have capacity to identify those high-risk patients, and could be used as a further reference to judge whether distant metastasis risk exists before treatment. However, the added prognostic performance could not be shown in the study. The AUC was 0.673 for the combination of radiomics features and risk clinical risk factors, and 0.640 for radionics features lonely. No significant difference between them (all of p-values >0.4, Delong test) was observed. Comparing to a previous study by Coroller et al.7 that demonstrated the c-index of 0.61, which was not significant different in the predictive performance of distant metastasis. We could not confirm that radiomics has complementary outstanding improvement on prediction of distant metastasis, or significant association with distant metastasis. In our exploration, three clinical factors and three radiomics features were selected, maybe the models based on pre-treatment images or parameters do most likely not contain enough information to accurately predict future event. This result might have not strong added predictive power, maybe we need to further re-analyse the pre-treatment images. Some patients underwent chemotherapy due to wider and bigger target volume before radiotherapy. In the future, it would be interesting to re-grouped the patients. Moreover, we need to further identify that the characteristics of the distribution that qualify feature as significant indicator differ from the property of the distribution that qualify predictive guess.24 It’s possible that radiomics features may be important for a particular outcome but may have not strong predictive power for the same outcome.
Prognostic prediction of overall survival
The prognostic values of the clinical risk factors and radiomics features were assessed according to the scoring system nomogram. Smoking status was prognostic (p-value< 0.01). Clinical stage IIB and IIIA were also prognostic (p-value< 0.01), and clinical stage IIIB had significantly prognostic (p-value< 0.001). In contrast, no significance was found on pathology (p-value> 0.05). Interesting, the above clinical risk factors become non-significant (p-value> 0.05) when rad_score signature was incorporated into the assessment model of nomogram (Figure1a). Perhaps the probable reason is that the stronger predictive power of radiomics features occupied that of clinical risk factors, thereby highlighting the potential value of radiomics in OS prediction of SCLC patients. The result is not surprising, as radiomics reflect higher imaging patterns and capture more imaging heterogeneity compared with clinical factors.
Radiomics describes intra-tumoral heterogeneity, might be related to the expression of genomic heterogeneity. These features would indicate worse prognosis than other factors, as tumors with more genomic heterogeneity were more likely to resistant to treatment and yield metastasis.4,25 The calibration of the two nomograms demonstrated that radiomics-based nomogram had a median C-index greater than that of only clinical risk factors-based nomogram, although the different was not significant.
In addition to the nomogram model, we also performed a subset analysis on the comparison between two KM curves. For obtaining the potential ability of radiomics on prediction of OS, all patient datasets were stratified into low- and high-risk groups according to the median of rad_score. The significant difference is observed in five-month OS. If no separating by radiomics features was done, it is likely that patients in the high-risk group of OS would be included into low-risk group and would miss necessary additional medical care. The incremental value of radiomics as a cut-off for precision OS prediction is well-demonstrated and successfully stratified patients into high- and low-groups (Log-rank, p-value < 0.0001).
The short-coming of this study was that the number of the patients with composite SCLC pathology was relatively small, and patients included in the study were only divided to two types of pathology, which maybe have influenced the generalization of the conclusions. Another limitation is that we could not consider the influence of respiratory motion on radiomics because four-dimensional CT in our institute is not available. It was shown that CT radiomics features can be susceptible to respiration.26 In our future work, composite SCLC patients should be further detailed and classified, according to more specific pathologies such as adenocarcinoma components or squamous cell carcinoma components or large cell carcinoma. Therefore, further prospective analyses including the various pathologies of SCLC patients will be needed to validate the prognostic significance of radiomics features.
In this study, nomogram based on radiomics features and clinical risk factors was proposed and estimated, and classifier model for predicting DM of patients with SCLC was developed. In addition, radiomics was as a complementary way to current prediction of DM and OS. Finally, a nomogram based on combining the radiomics signature and clinical factors provided a convenient way to assess OS for SCLC patients. The prediction can be used to make recommendations for improving precision decision-making.
The authors declare that they have no competing interests.
Ethics approval and consent to participate: This study with clinical data was approved by the Tianjin Cancer Hospital. Medical Ethics Committee and informed consent was waived.
This work was supported by the Natural Science Foundation of Tianjin, China (20JCYBJC01510), and National Science Foundation of China, China (11875171, 81872472).

©2022 Zhang. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.