Journal of
eISSN: 2373-4469


Research Article Volume 4 Issue 1
1Bioinformatics Department, Sadat City University, Sadat City, Egypt
2Biology Department, King Abdulaziz University, Saudi Arabia
3Botany Department, Kafrelsheikh University, Egypt
4IME, Fraunhofer Institute, Germany
Correspondence: Haddad A El Rabey, Bioinformatics Department, Genetic Engineering and Biotechnology Institute,Sadat City University, Sadat City, Minufiya, Egypt
Received: November 09, 2016 | Published: January 26, 2017
Citation: El Rabey HA, Albureikan MO, Aly MM, et al. Isolation, cloning and sequencing of poly 3-hydroxybutyrate synthesis genes from local strain of bacillus cereus mm7 and expressing them in E. Coli. J Investig Genomics. 2017;4(1):7-14 DOI: 10.15406/jig.2017.04.00056
Poly 3-hydroxybutyrate (PHB) is the most known degradable biopolymers, produced by some genera of bacteria under unfavorable growth conditions. Isolation, cloning, and expression of PHB genes from PHB producing Bacillus cereus was achieved. A highest Poly 3-hydroxybutyrate producer bacterium was selected and identified as Bacillus cereus MM7, based on morphological and physiological characters in addition to 16S rRNA gene sequences. The four genes of Bacillus cereus MM7 that involved in Poly 3-hydroxybutyrate production were successfully amplified using specific primers which were designed depending on the sequence alignment of the known PHA biosynthetic genes from Bacillus cereus, cloned in E. coli DH5α and completely sequenced. Moreover, the expression of the four genes responsible for PHA biosynthetic in E. coli XL1-Blue and E coli MJ 109 was assayed by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis. In the current study Bacillus cereus was able to synthesize PHB. PHB genes were isolated, cloned and expressed in E. coli DH5α. The PHB encoding proteins were analyzed in SDS-PAGE and the PHB corresponding protein bands were analyzed using mass spectrometry to nominate their amino acid sequence.
Keywords: Bioplastic; Bacillus; PHB; Gene; Cloning; Expression; Polyacrylamide
Poly 3-hydroxybutyrate (PHB) is a polyhydroxyalkanoate (PHA) which is a polymer belonging to the polyesters class that are of interest as bio-derived and biodegradable plastics produced by some genera of bacteria under unfavorable conditions [1].
PHA can be accumulated by microorganisms in the environment naturally or PHA can be produced artificially by microorganisms if they are fed with specific chemicals in the laboratory which are not the normal substrates for cell growth and survival [2]. It is the only bioplastics completely synthesized by over 30 % of soil bacteria [3].
PHA synthases are divided into four groups i.e. I- II- III and IV, based on the enzymes substrate specificities and the enzymes subunit composition [4]. The three classes I (found in Ralstonia eutroPHA), III (found in Allochromatium vinosum), and IV (found in Bacillus megaterium) PHA synthases have substrate specificities toward short-chain-length monomers (C3-C5), whereas class II (found in Pseudomonas aeruginosa) PHA synthase utilizes medium-chain-length monomers (C6-C14) [5].
Excess carbon sources with limitation of nitrogen and phosPHAte were used to produce PHB in different kinds of bacteria [6,7]. Furthermore, Ghate et al. [8] isolated different Bacillus species for the production of polyhydroxybutyrate (PHB) from soil samples. Sangkharak et al. [9] isolated twenty five Bacillus sp. strains that were able to accumulate polyhydroxyalkanoates (PHAs).
Davis et al. [10] used new pathway employing genes for b-ketothiolase and NADPH dependent acetoacetyl CoA reductase (PHAA and PHAB) isolated from Bacillus sp. 256 and genes for PHA synthase and (R)-specific enoyl CoA hydratase (PHAC1 and PHAJ1) cloned from P. aeruginosa for the production of PHB in recombinant E. coli. PHA biosynthetic genes from different species were cloned for mass production of PHAs for commercial use [6,11-13]. Tomizawa et al. [14] studied the expression of PHA synthases from Bacillus megaterium NBRC15308 (PHARCBm), B. cereus YB-4 (PHARCYB4) and hybrids (PHARBmCYB4 and PHARYB4CBm) in Escherichia coli JM109 to characterize the molecular weight of the synthesized poly (3-hydroxybutyrate) [P (3HB)].
The aim of the current study was the selection and identification of the most active PHB producing bacteria, isolating the PHB genes, cloning and sequencing them and assaying the expression of PHB genes in Escherichia coli. The PHB encoding proteins were analyzed in SDS-PAGE and the PHB corresponding protein bands were analyzed using mass spectrometry to nominate their amino acid sequence.
A highly PHB producing isolate was isolated in our lab from sea water sample collected from North Beach of Jeddah, Saudi Arabia and identified as Bacillus cereus MM7 based on morphological and physiological characters in addition to 16S rRNA gene sequences [15]. This bacterium was used as a source for PHB genes.
Pre culture
The pre culture was prepared in 250 ml Erlenmeyer flasks containing 50 ml of nutrient broth [16]. Each flask was inoculated with a loop of the tested cultures and incubated in a 130 rpm speed rotary shaker at 30˚C for 24h.
Isolation of genomic DNA
The DNA of PHB producing strain was extracted using Nucleo Spin Tissue Kit (Macheri Nagal, Germany) according to the manufacturer’s instruction.
Colony PCR
For colony PCR amplification, bacterial colonies were picked using a sterile tooth pick and dipped into 10µl of water plus 10µl PCR reaction mixture consisting of 0.4µl of forward and 0.4µl reverse primers, 0.5µl dNTPs, 2 µl PCR buffer, 0.6µl MgCl2, 0.3µl Taq DNA polymerase and 5.8 µl. Amplification reaction was performed using MWG-Biotech Primus 96 thermocycler, Galileo Equipos S.L, Spain) under the following reaction conditions; initial denaturation at 95˚C for 5 min, followed by 25 cycles denaturation at 94˚C for 1m, annealing at 55˚C for 1m and elongation at 74˚C for 3 min and final elongation at 72˚C for 10 min.
Recombinant E. coli medium
The medium used for E. coli growth was LB medium (5 g yeast extract, 10 g tryptone, 10 g NaCl per liter of deionized water). For PHB production, it was supplemented with 20 g/L glucose in a 500-ml shaking flask containing 100-ml medium at 37˚C for 24h. Sterilized ampicillin was added to the E. coli culture medium to a final concentration of 100μg/ml.
PCR amplification of the PHA genes
The isolated DNA from B. cereus MM7 strain was used as a template for PCR amplification of PHA biosynthesis genes which contain (PHAA, PHAR, PHAB and PHAC). The oligonucleotide primers for PCR amplification are listed in Table 1. The primer of PHAA gene was designed based on the sequence alignment of the known PHA synthases from the B. cereus group (organisms B. cereus, accession number EU239690) [10], while the oligonucleotide primers for PCR amplification of PHAR, PHAB and PHAC were designed in this study based on the sequence alignment of the known PHA synthases from B. cereus. (Accession number AB525763) [17].
Strains/ Plasmid / Primers |
Relevant Description |
Source or |
E. coli DH5α |
Cloning strain |
Biolab. |
E. coli XL1-Blue |
Expression strain |
promega |
E. coli JM109 |
Expression strain |
promega |
pJET1.2/blunt cloning vector |
linearized with Eco32I (EcoRV) (GenBank/EMBL Accession number EF694056), and has the highest efficiency cloning of PCR products |
Thermo Scientific CloneJET PCR Cloning Kit. |
Fw pJET1.2 5’-d(CGACTCACTATAGGGAGAGCGGC)-3’ |
pJET1.2 primers |
From Thermo Scientific Fermentas™ pJET™1.2 |
Rv pJET1.2 5’-d(AAGAACATCGATTTTCCATGGCAG)-3’ |
||
Fw (1) PHAA 5-TACGTAACTAATAACGAGGGGAG-3 |
Amplification of PHAA gene of Bacillus cereus for cloning it into pJET |
Designed at |
Rv (1) PHAA 5-GCTTCCAACGCCCTCCTGTTCTAAACG-3 |
||
Fw (2) PHAA 5-CTAATAACGAGGGGAGATATCGTTC-3 |
||
Rv (2) PHAA 5-GTCGTTTCACCCCTTCCTGTGAGGC-3 |
||
Fw (1) PHARBC operon 5-AAATATAGACTTGAAGTTTTATTCAG-3 |
Amplification of PHARBC genes of Bacillus cereus for cloning it into pJET |
At |
Rv (1) PHARBC operon 5-AACATAGGCTAGTTGGATTTTTTAT-3 |
||
Fw (2) PHARBC operon 5-GACTTGAAGTTTTATTCAGAATATTAG-3 |
||
Rv (2) PHARBC operon 5-CATAAAAAAATCCAACTAACATAGGC-3 |
||
Fw PHARBC operon: 5- GCGCTTGTTTATAACATATGAAAG-3 |
Amplification to detection of B gene of Bacillus cereus |
Designed at |
Rv PHARBC operon: 5-CTATCTCCTTTTTGGTCAATTTC-3 |
||
Fw PHAA 5-ATTAATAATGCGGCCGCCATGAGAGAAGCTGTCATTG-3 |
Amplified PHAA gene for subcloning it into pJET-PHARBC(+) |
Designed at |
Rv PHAA 5-ATTAATAATCTCGAGTTATAGTAATTCAAATACTCCCG-3 |
Table 1: E. coli strains, plasmid and primers used in this study.
PCR amplification protocol for PHAA, PHAB, PHAC and PHAR
PCR reactions were achieved using genomic DNA as a template in 25µl PCR reaction mixture consisted of 2.5 µl PCR buffer, 0.5µl of each forward and reverse primers, 0.5µl dNTPs, 0.5µl polymex enzymes, 1µl DNA (containing 50-100 ng DNA) and 19.5µl water. PCR reaction condition was as follows: under the following reaction conditions; initial denaturation at 95˚C for 5 min, followed by 25 cycles denaturation at 94˚C for 1m, annealing at 55˚C for 1m and elongation at 74˚C for 3 min and final elongation at 72˚C for 10 min. The PCR product was excised using clean scalper under UV light, purified using NucleoSpin® Gel and PCR Clean-up kit (Macheri Nagal, Germany) and then sequenced (at DNA sequencing unit at Fraunhofer Institute for Molecular Biology and Applied Ecology, Germany).
The PCR product was resolved on 0.8% or 1.2% agarose gel (w/v agarose in 1X TBE) buffer depending on the size of DNA and the experiment. The running buffer was 1X TBE buffer. The gel was stained with 0.1μg ml ethidium bromide, visualized under UV light, and then photographed using gel documentation system.
Ligation of PHAA PCR product to pJET-PHARBC
The ligation was performed according to the instruction of ligation protocol supplied with T4 DNA ligase (New England Biolabs).
Bacterial transformation of DH5α, E. coli XL1-blue and E. coli JM109
Heat-shock transformation was performed in DH5α, E. coli XL1-Blue and E. coli JM109 according to the instruction of Lamitina Lab Protocols.
Cloning of PHA synthase genes from the strain Bacillus cereus MM 7
All E. coli strains, plasmid and primers are listed in Table 1. The purified PCR product was ligated to the pJET1.2/blunt cloning vector (Clone JET PCR Cloning Kit, Fermentas, Thermo Fisher Scientific Inc) according to the instructions of the supplier to get three kinds of plasmids; pJET-PHAA (Figure 1A), pJET-PHARBC (Figure 1B) and pJET-PHAARBC (Figure 1C). After plasmid was extracted using miniprep Kit (Nucleo Spin Plasmid kit, Macheri Nagal, Germany). All the three types of plasmids were transformed separately into E. coli DH5α using heat shock transformation protocol according to Lamitina Lab protocols. Afterwards, the plasmid was extracted and the concentration of plasmid was measured and digested with XhoI (Biolabs, England) to check the size after cutting with restriction enzyme and check if there is other site can be cut. Oligonucleotide primers containing NotI and XhoI restriction sites sequences were designed by Biolabs for PCR amplification of PHAA gene in pJET +PHAA. After that, the PCR product of PHAA and (+) pJET +PHARBC were digested with (NotI and XhoI), and then PHARBC and PHAA were ligated to each other and to the pJET vector according to the ligation protocol of T4 DNA ligase protocol (M0202), (New England Biolabs) to get (+) pJET +PHAARBC operon (Figure 1C). The construct was transformed into E. coli DH5α, E. coli XL1-Blue and E. coli MJ 109. The (+) pJET +PHAARBC operon was sequenced at (Fraunhofer Institute for Molecular Biology and Applied Ecology, Germany).
Determination of the (+) orientation for pJET +PHARBC operon and (+ pJET+PHAARBC using HindIII restriction enzyme
The PHARBC operon was then digested with HindIII to determine the orientation, if it is (+) or (-) before ligation and also to determine the orientation for PHAARBC if it is (+) or (-). The samples were resolved on agarose gel, and then estimated the bands size to know the kind of orientation.
Determination of the (+) orientation for pJET +PHARBC operon and (+) pJE+PHAARBC by colony PCR
To determine the orientation of pJET +PHARBC operon, if it is (+) or (-), colony PCR amplification was achieved using forward B gene detection primer and forward pJET1.2 primer (which must give no band in the gel with only (+) orientation for pJET +PHARBC operon). Also, it was done using forward B gene detection primer and the reverse pJET1.2 primer (which must give band in the gel with only (+) orientation for pJET +PHARBC operon). After detecting the positive colony was incubated in 5 ml LB medium and the plasmid was isolated and its concentration was measured using Nano drop spectrophotometer. PCR was run using the same primers that was used in colony PCR to assure results. On the other hand, to check the orientation for pJET +PHAARBC if it is (+) or (-) after ligation, forward pJET1.2 primer and reverse PHAA gene primer were used for amplification (It must give band in the gel with (+) orientation). Then the positive colony was inoculated in 5 ml LB medium and the plasmid was isolated and its concentration was measured using Nano drop spectrophotometer. PCR amplification was achieved using the same primers listed in Table 1 and the same PCR reaction conditions mentioned above.
PHB genes expression
The gene expression of PHB genes was achieved according to McCool et al. [18]. After getting the (+) pJET - PHAARBC operon construct, it was transformed to E. coli DH5α, E. coli XL1-Blue and E. coli MJ 109, and then grown overnight.
The transformed strains were grown at 37˚C in Luria Bertani medium alone and also in Luria Bertani medium supplemented with glucose. The crude extracts of E. coli strains were prepared after 7h and 24h of incubated by sonication. Then, spun for 5 min at 11000 rpm. The crude extracts of E. coli strains were precipitated and soluble proteins (supernatant) were separated from insoluble proteins (pellet). Then, proteins were loaded on SDS-PAGE 12% (w/v) polyacrylamide gel and run at 35mA for 1hour.
All the expected protein bands of PHB genes expression (PHAA 41,13 KD: PHAB 26.54KD: PHAC 41.69KD: PHAR 18.5KD ) were excised from the SDS-gel and analyzed using mass spectrometry (Mascot software. Matrix science) at the proteins analyses unit in Fraunhofer Institute for Molecular Biology and Applied Ecology, Germany. The amino acid sequence of all PHB genes was compared to that present in the databases using BLAST search to find the most homologous sequences to them.
PCR amplification of the PHB genes
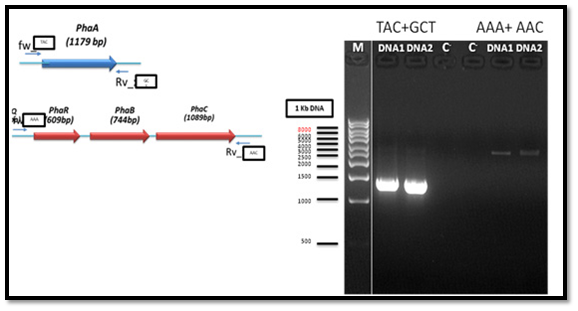
The genomic DNA of PHA producing bacteria (Bacillus cereus MM7) was used as a template for PCR amplification of the PHA biosynthesis genes which consists of PHAA, PHAR, PHAB and PHAC. Two of the oligonucleotide primers for PCR amplification of PHAA gene gave positive result with clear bands in the gel while the other two primers did not give bands. Also, the oligonucleotide primers for PCR amplification of PHAR, PHAB and PHAC operon gave positive result with clear bands in the gel, whereas the other two primers did not give any band. PCR reaction resulted in a successful amplification of an approximately 900 bp (for partial fragment) and distinct 1.8 kb (for full-length gene) from the tested bacteria as shown in Figure 2.
Cloning of PHA synthesis genes from the strain Bacillus cereus MM7
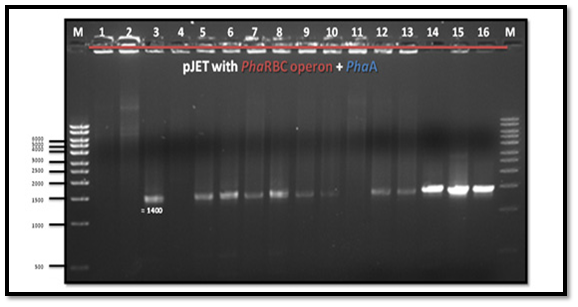
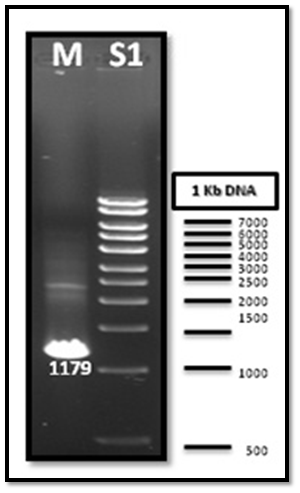
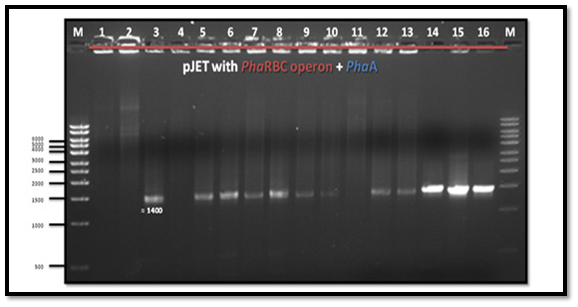
After PCR product purification to PHAA and PHAR, PHAB, PHAC operon and cloning them into the pJET1.2/blunt cloning vector, three recombinant plasmids (pJET-PHAA, pJET-PHARBC operon and pJET-control) were obtained. These plasmids were successfully transformed separately into E. coli DH5α by using heat shock transformation. Then, again the plasmid was extracted and digested with XhoI to check the size after cutting with restriction enzyme and check if there is another sites can be cut and the results showed that the size of pJET-PHAA (4221pb) and for pJET-PHARBC operon (5809pb) were correct as expected and it was also appeared that only one restriction site in the plasmid was there, which mean that all the four genes PHAA and PHAR, PHAB, PHAC operon were successfully cloned in two pJET1.2/blunt cloning vectors (Figure 3). Then, the PHAA gene in pJET +PHAA was amplified and digested with restriction enzymes NotI and XhoI before running the digestion product in the gel to check the size of PHAA gene (1179pb) (Figure 4). The (+) pJET +PHARBC operon with NotI and XhoI was digested and successfully ligated to get (+) pJET +PHAARBC operon which transformed to E. coli DH5α, E. coli XL1-Blue and E. coli MJ 109.
Detecting if the pJET+PHAA was successfully ligated to pJET+PHARBC operon
The forward primer pJET1.2 and the reverse primer PHAA were used to detect the orientation of pJET +PHAARBC, whether it is (+) or (-). The PCR result showed a band which means that pJET +PHAARBC was (+) orientation (Figure 5).
The sequence data of PHAA
The sequence data of PHAA was 1186 bp as shown in the Supplementary Figure 1. The BLAST search of this sequence showed 99% homology to the sequence of acetyl-CoA acetyltransferase from Bacillus thuringiensis BMB171, accession No. (NC_014171.1), [19] and Bacillus cereus ATCC 14579, accession No.: NC_004722.1 [20].
The sequence data of PHAB
The sequence data of PHAB fragment is 745 bp as shown in the Supplementary Figure 2 and the BLAST search showed 98% homology to acetoacetyl-CoA reductase of Bacillus thuringiensis serovar finitimus YBT-020 NC_017200.1 [21] and Bacillus thuringiensis serovar finitimus YBT-020 NC_017200.1 [21] and Bacillus cereus FRI-35, accession No.: NC_018491.1.
The sequence data of PHAC
The sequence data of PHAC is 1086 bp as given in the Supplementary Figure 3 and the BLAST search showed 98% homology to poly(R)-hydroxyalkanoic acid synthase subunit PHAC from Bacillus thuringiensis serovar finitimus YBT-020, accession No. NC_017200.1 [21].
The sequence data of PHAR
The sequence data of PHAR is 484 bp as given in Supplementary Figure 4 and also aligned sequence ata of PHAR using BALAST search, showed 99% homology to PHA synthase subunit PHAR; regulator of PHAP of Bacillus cereus ATCC 10987, accession No.: NC_003909.8 [22].
The complete sequence of the PHA operon
The complete sequence of the PHA operon is 4279 bp as shown in Supplementary Figure 5. The BLAST search gave 98% homology to PHAR protein acetoacetyl-CoA reductase of Bacillus cereus ATCC 14579, accession No. NC_004722.1 [20].
PHA synthase gene expression
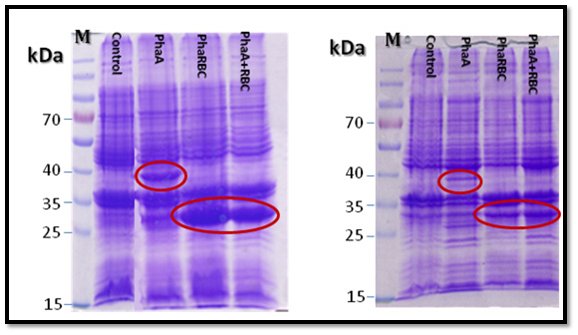
All the expected protein bands in the SDS-gel (PHAA 41,13 KD: PHAB 26.54KD: PHAC 41.69KD: PHAR 18.5KD ) were cut and the proteins were analyzed using mass spectrometry (Mascot software, Matrix science) at the proteins analyses unit in Fraunhofer Institute for Molecular Biology and Applied Ecology, Germany. The result confirmed the presence of PHAA, PHAR and PHAB, which means that PHAA, PHAR and PHAB are expressed. But there was no clear protein band in the gel for PHAC which means that PHAC is probably expressed with very little amounts or not expressed. Also, the current results showed that using the proteins pellet better than soluble proteins in SDS-PAGE gel for expression analysis (Figure 6).
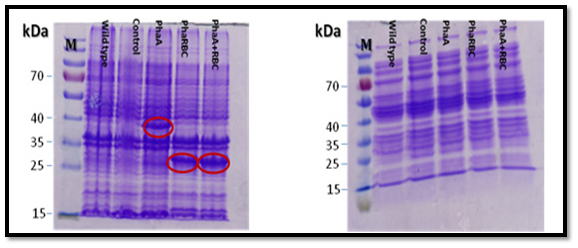
Moreover, the proteins were expressed clearly when using medium not supplemented with glucose better than using medium supplemented with glucose (Figure 7). The proteins bands were clearer when the strains were incubated for 24 h than 7 h as shown in Figure 8.
PHA expression
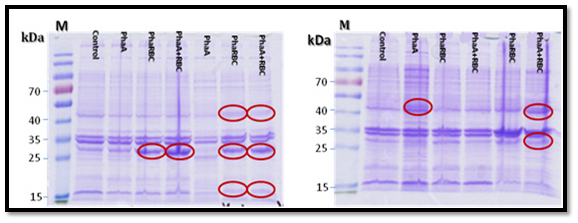
The expression of PHA genes in E. coli (transformed with PHA genes), showed the production of three protein subunits, PHAA, PHAB, PHAR and the absence of PHAC protein subunit. The MS analysis of the three proteins showed that PHA was 390 amino acids as shown in the supplementary materials figure 6, PHAB was 247 amino acids (Supplementary Figure 7) and the PHR was 176 amino acids (Supplementary Figure 8). On the other hand, the BLASTP search of the amino acid sequence for PHAA subunit. The PHAA amino acid sequence showed 100% homology to acetyl-CoA acetyltransferase ATCC 14579 of Bacillus cereus Accession No.: NP_834674.1 [20].
Four classes of PHA synthase have been classified based on subunit composition of enzymes and their substrate specificities [23]. Bacillus strains belong to class IV synthase that comprises PHAC and its subunit PHAR [24]. The most important genes that involves in PHB production in Bacillus strains were PHAA and PHAR, PHAB, PHAC [3,23]. On the other hand, Valappil et al. [25] and Tomizawa et al. [14] proved that in Bacillus genus, two proteins, PHAC, the normal PHA synthase enzyme, and PHAR have been proven to be essential for the PHA synthase activity. The PHA synthases was exist in two forms; an active form in PHA-accumulating cells and an inactive form in non-accumulating cells, where PHAC is more susceptible to degradation in cell [18].
All PHA genes; PHAA, PHAR, PHAB, PHAC were amplified and assembled together, and then cloned in E. coli. The results showed that PHAA sequence (1186 bp) gave the maximum homology of 99% to acetyl-CoA acetyltransferase of Bacillus thuringiensis BMB171, accession No.: NC_014171.1 (He et al. 2010) and Bacillus cereus ATCC 14579, accession No.: NC_004722.1 [20]. In addition, the sequence data of PHAB fragment was found 745 bp and the BLAST search gave a maximum homology of 98% to many Bacilli strains sequence e.g. acetoacetyl-CoA reductase of Bacillus thuringiensis serovar finitimus YBT-020 NC_017200.1 [21] and Bacillus cereus FRI-35, accession No.: NC_018491.1. Moreover, the sequence data of PHAC was found 1086 bp and the BLAST search of PHAC sequence showed 98% homology to poly(R)-hydroxyalkanoic acid synthase subunit PHAC from Bacillus thuringiensis serovar finitimus YBT-020, accession No. NC_017200.1 [21]. Also, the sequence data of PHAR was 484 bp and the BLAST search of homologous sequence to PHAR showed 99% homology to PHA synthase subunit PHAR; regulator of PHAP of Bacillus cereus ATCC 10987, accession No.: NC_003909.8 [22] and Bacillus thuringiensis serovar finitimus YBT-020, accession No.: NC_017200.1. Finally, the complete sequence of the PHA operon resulted from PCR amplification and sequencing using the specific primers was 4279 bp and the BLAST search gave 98% homology to PHAR proteinacetoacetyl-CoA reductase of Bacillus cereus ATCC 14579, accession No. NC_004722.1 [20]. This result is consistent with many studies in different bacteria such as; Rhodospirillum rubrum [26], Bacillus megaterium [18], Bacillus cereus SPV [25] and Bacillus thuringiensis R1 [4].
It is worthy to mention that in this study, it is the first time to isolate and clone all the four genes; PHAA and PHAR, PHAB, PHAC responsible for PHB production from the same bacterial isolate “Bacillus cereus MM7”, in contrast with the previous studies which used only two or three genes of PHB biosynthetic genes and cloned them in plasmid that carry the remaining PHB biosynthetic gene [4].
The four PHA genes were cloned and expressed in E. coli XL1-Blue and E. coli MJ 109 and the resulting proteins were analyzed using SDS-PAGE [18]. The results of polyacrylamide gel electrophoresis and proteins mass spectrometry data (Mascot software. Matrix science) confirmed that the PHAA (41,13 KD), PHAR (18.5KD) and PHAB (26.54KD) were expressed very well and the their bands in the gel were also clear, but there was no clear protein band in the gel for PHAC (41.69KD) which means that PHAC is probably expressed with very little amounts or not expressed. The MS analysis was performed for the corresponding protein bands for PHB genes according to Slater et al. [27], Valappil et al. [25] and Zhu et al. [13].
The current results showed that using the proteins pellet better than soluble proteins in SDS-PAGE gel to analyze the expression, this may be due to the elongation of PHA chain and PHAsing proteins or the specific proteins that are associated with the produced PHA granules or budding of a vesicle with a phospholipid layer and leading to granule formation [2]. Many other studies used whole-cell extracts for determination of one protein or more of PHA synthesis proteins activity [10,18,28]. In contrast, Zhu et al. [13] used soluble protein to detect PHB expression proteins (PHBA, PHBB and PHBC) which were all expressed at high levels. Moreover, in the current study, the PHB encoding proteins were expressed clearly when using a medium not supplemented with glucose better than using glucose. This result agrees with Davis et al. [10] who used LB media without glucose and noted that PHAC1, PHAJ1, PHAA and PHAB proteins were expressed very clear.
In the current study, the amino acid sequence analysis of the three proteins using MS showed that PHAA was 390 Amino acids with 100% homology to acetyl-CoA acetyltransferase ATCC 14579 of Bacillus cereus Accession No.: NP_834674.1 [20]. PHAB was 247 amino acids and it showed 100% homology with acetoacetyl-CoA reductase (Bacillus cereus ATCC 14579) accession No.: NP_831099.1 [20]. PHAR was 176 amino acids and it showed 89% similarity to hypothetical protein [Bacillus cereus], accession No.: WP_002195298.1. Similar results were obtained in many previous studies which sequenced and analyzed the amino acids of PHB proteins in Rhodospirillum rubrum [26], Pseudomonas sp. [29,30] Bacillus sp. [4,18,25].
In conclusion, Bacillus cereus MM7 isolate was able to synthesize Poly 3-hydroxybutyrate. The PHB genes were isolated, cloned, sequenced and expressed in E. coli DH5α. The PHB encoding proteins were analyzed in SDS-PAGE and the PHB corresponding protein bands were analyzed using mass spectrometry to nominate their amino acid sequence.
The authors thank King Abdulaziz City for Science and Technology for financing this research project grant number (1013-12-ت-ط). Thanks also for Fraunhofer Institute for Molecular Biology and Applied Ecology, Germany for awarding the facilities needed to perform this research project.

©2017 El, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.