Journal of
eISSN: 2373-4469


Research Article Volume 2 Issue 2
1Department of Genetics, Sanjay Gandhi Post Graduate Institute of Medical Sciences, India
2Department of Neurology, Sanjay Gandhi Post Graduate Institute of Medical Sciences, India
Correspondence: Sarita Agarwal, Department of Genetics, Sanjay Gandhi Post Graduate Institute of Medical Sciences, Lucknow, India- 226014, Tel 91 522 249 4349, Fax 9152 2266 8017
Received: January 12, 2015 | Published: March 4, 2015
Citation: Kumar A, Agarwal S Pradhan S. GST polymorphisms and GST enzyme activity in type 1 myotonic dystrophy. J Investig Genomics. 2015;2(2):33-41. DOI: 10.15406/jig.2015.02.00020
Glutathione S-transferases (GSTs) play an important role in protecting cells from damage caused by endogenous and exogenous compounds. In present study, we investigated the association of GST gene polymorphisms (GSTT1, GSTM1, GSTP1 and GSTM3) and its enzyme activity in DM1 affected Indian population. Serum GST level was assessed by using GST-kit. GSTM1 (null or present) & GSTT1 (null or present) GSTM3 (AA, AB and BB) & GSTP1 (Ile/Ile, Ile/Val and Val/Val -105) polymorphism were analyzed. The serum GST significantly reduced in the patient group compared to the control group and significantly correlated with diabetes. Patients had significantly higher (except GSTM3A/B) GSTM1*0 (GSTM1 null genotype), GSTT1*0 and GSTP1 (ile/val) frequency than controls. The deletion frequencies (GSTM1 and GSTT1) and GSTP1 (ile/val) were not associated with higher risk while heterozygous frequency of GSTM3 (A/B) increased risk ratio up to three fold. The GSTM1, GSTT1, GSTM3 genotypes correlated with dyspepsia, age at presentation and duration of disease respectively. The allelic frequencies were 0.46, 0.54 and 0.48, 0.52 for GSTM3*A and GSTM3*B and were 0.61, 0.39 and 0.55, 0.45 for ile (A) and val (G) of GSTP1 for DM1 patients and control group respectively. The group of combination genotype frequency had no impact on higher risk of disease. GSTT1 and GSTM1 seem to be a candidate gene for susceptibility to DM1 in Indian population. Further studies using genetic polymorphisms of glutathione or other antioxidant enzymes are required to clarify the relationship between increased oxidative stress and DM1.
Keywords: myotonic dystrophy type 1 (dm1), polymorphism and activity, gstm1, gstt1, gstm3, gstp1
GSTs, glutathione s-transferases; DM, myotonic dystrophy; PROMM, proximal myotonic myopathy; ROS, reactive oxygen species; Ile, isoleucine; Val, valine; EMG, electromyographia; NCV, nerve conduction velocity; OD, optical density; SNP, single nucleotide polymorphisms; LWD, learning and writing disability; SLD, speech and languages disability; SCK, serum creatine kinase; CK-MM, creatine kinase-muscle isoform; CK-MB, creatine kinase-isoform; PCR, polymerase chain reaction; +/+/+, gstm1, gst1 and gstm3 present; -/-/-, gstm1, gstt1 and gstm3 absent; ile/val and ile/ile=heterozygous and homozygous status of GSTP1
Myotonic dystrophy (DM) is an autosomal dominant, multisystem trinucleotide repeat disorder. Myotonic dystrophy is clinically heterogeneous and at the molecular level at least two types can be distinguished: DM type 1 (DM1; Steinert disease) and DM type 2 (DM2; proximal myotonic myopathy (PROMM) or Ricker syndrome). DM1 is the most common form of muscular dystrophy in adults with an estimated incidence of 1:8000.1 Two different mutations are responsible for DM: DM1 (OMIM #160900) is caused by a (CTG)n repeat expansion in the 3’-untranslated region of the DMPK gene located within chromosome band 19ql3.32,3 while DM2 (OMIM #602688) is caused by a large (CCTG)n repeat expansion in intron l of the CNBP gene at chromosome 3q21.4,5 GSTs activity involves in the pathogenesis of DM1 because it contains polymorphic trinucleotide repeat.6 Human GSTs represent a large and diverse super family of enzymes, with at least 13 GST enzymes belonging to five different families: mu, theta, alpha, pi, and gamma. The GSTM1 & GSTM3, GSTT1 and GSTP1 belong to the GSTmu, GSTtheta, and GSTpi categories of enzymes respectively. Glutathione S-transferases (GSTs) are phase II detoxifying enzymes that catalyze a variety of reduced glutathione-dependent reactions with electrophilic substrates, including active metabolites of carcinogens.7 In particular, GSTM1, GSTM3, GSTP1 and GSTT1 are involved in detoxification of active metabolites of several potential carcinogens such as benzo[a]pyrene and other polycyclic aromatic hydrocarbons, monohalomethanes and ethylene oxide.7‒9 It also play an important role in the functioning of antioxidant defences through reactive oxygen species (ROS) metabolism, in the repairing of damaged ROS and in the detoxification of several xenobiotics.7,10
The absence of GSTT1 and GSTM1 enzyme activities is caused by homozygous deletion of the respective genes (null genotypes).11,12 In the GSTM3 gene, the GSTM3*A wild-type allele and the GSTM3*B variant allele have been described.8 GSTM3*B contains a recognition motif for the YY1 transcription factor, which has been postulated to regulate gene expression.13 It was suggested that GSTM3*A and GSTM3*B are expressed at different levels and could therefore confer different efficiencies in the metabolism of carcinogens and toxic compounds.14 In the GSTP1 gene, 2 variant alleles, GSTP1*B and GSTP1*C, have been detected in addition to the wild-type allele GSTP1*A.9 Both variant alleles have an A-to-G transition at nucleotide 313, causing a change of isoleucine (Ile) to valine (Val) at codon 105. The specific activity and affinity for electrophilic substrates is altered in the Val variant.9 It is therefore plausible that hereditary differences in the activities of these enzymes, due to genetic polymorphisms.8,9,11,12
Different GSTs polymorphism has been studied in various malignancies.15‒18 However, there are no data on GST polymorphism and DM1 from India and abroad. This highlights the need for investigation of the association between GST polymorphism and the risk of DM1 in the population. Therefore, the aim of the present study was to investigate the association of GST gene polymorphisms (GSTT1, GSTM1, GSTP1 and GSTM3) and its enzyme activity in DM1 affected Indian population.
Diagnosis and collection of samples from dm1 patients and control subjects
Patients having complaint of muscle wasting, jaw and temporal wasting, impairment in gripping capacity, arrhythmia, facial weakness and hypersomnia included in the study while patients having any other neurological disorder or any other severe or familiar disease excluded from the study. Clinically, DM1 patients diagnosed on the basis of detailed history of disease, physical examination, biochemical testing and electrophysiological testing like NCV (Nerve conduction velocity), EMG [Electromyographia, (Figure S1) etc. while molecular testing was done in all patients by TP-PCR methodology19 (Figure S2). Blood samples were collected in EDTA and plain vials from twenty-seven DM1 patients (median age 32.8years±9.3, range 17–52) and hundred age and sex matched controls (median age 31.0years±8.6, range 16–54) after written informed consent.
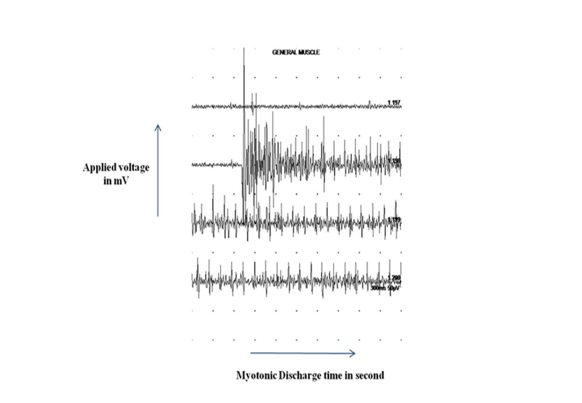
Figure S1 Concentric needle electromyography showing a typical myotonic discharges (20 second) in a myotonic dystrophy proband (III-4) with variation in amplitude as well as frequency, triggered by mechanical stimulation of abductor pollicis brevis muscle.
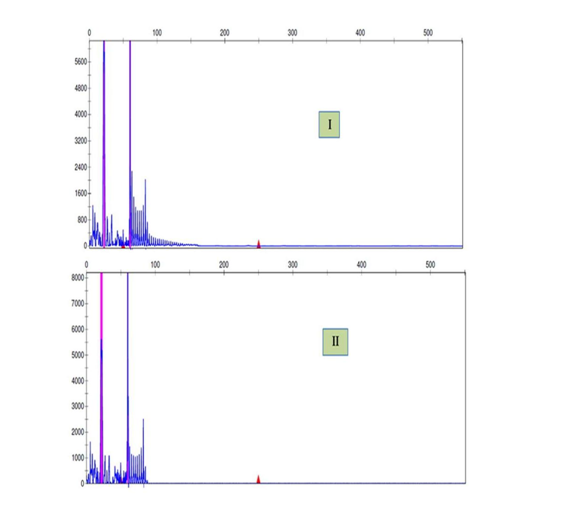
Figure S2 TP-PCR diagnosis of the myotonic dystrophy type 1.
The TP-PCR electropherogram represent the pathogenic CTG repeat expansion in a patient (I) and normal pattern in his mother (II).
Isolation, quantification and storage of DNA
Blood drawn in EDTA vials were processed for DNA isolation by standard phenol chloroform method. The quality and purity of DNA was checked by measuring optical density (OD) at 260nm and 280nm. The ratio of absorbance at 260 and 280 nm of DNA was around 1.7-1.9. The quality and purity was confirmed by 0.8% agarose gel electrophoresis in 1X TBE buffer and stored at -20 ͦ C till further use.
Serum isolation and analysis of GST activity
Blood sample collected in plain vials were centrifuged at 3,000g for 10 min at 4 ͦC; top layer were separated and stored at -80 ͦ C till analysis. GST level was assessed by using kit (Glutathine-S-Transferase assay kit, 96 wells, Cat. No. 703302-96) of Cayman Chemical Company 1180 East Ellsworth Road Ann Arbor, Michigan 48108, USA.
Genotyping
The samples collected from patients and controls were studied for single nucleotide polymorphisms (SNP) of the following genes: GSTMI (null or present), GSTT1 (null or present), GSTP1 (Ile/Ile, Ile/Val and Val/Val -105) and GSTM3 (AA, AB and BB). Primer sequences and restriction enzyme8,20,21 used in the study for different polymorphisms are given in Supplementary Table S1.
Name |
Primer Sequence |
Method |
Enzyme |
Product size |
|---|---|---|---|---|
GSTM1 |
F5’- TTCCTCACTGGTCCTCACATCTC-3’ |
PCR |
- |
Null-215bp |
R5’- TCACCGGATCATGGCCAGCA-3’ |
||||
GSTT1 |
F5’- GAACTCCCTGAAAAGCTAAAGC-3’ |
PCR |
- |
Null-480bp |
R5’- GTTGGGCT CAAA TATACGGTGG-3’ |
||||
GSTM3 |
F: 5'-CCTCAGTACTTGGAAGAGCT-3' |
PCR-RFLP |
MnlI |
11bp, 51bp, |
R: 5'-CACATGAAAGCCTTCAGGTT-3' |
86bp,125bp,134bp |
|||
GSTP1 |
5´- ACCCCAGGGCTCTATGGGAA -3' |
PCR-RFLP |
Alw261 |
176bp, 91bp, |
5'- TGAGGGCACAAGAAGCCCCT -3' |
85bp |
Table S1 Primer sequences used for polymorphism study
GSTM1 and GSTT1 polymorphism: Polymorphism was genotyped using specific primer sets for the GSTM1 and GSTT1 genes. The PCR mixture (25μL) for GSTM1 and GSTT1 genotyping contained 15µl of master mix, 15 pmol of each forward and reverse primer, 3µl of PCR water and 100ng of genomic DNA was used as template. The PCR conditions were as follows: initial denaturation at 94 ͦC for 2min, 35 cycles of denaturation at 94 ͦC for 1min, annealing at 55 ͦC and 62 ͦC respectively for GSTM1 and GSTT1 for 1min, extension at 72 ͦC for 1min, and a final extension at 72 ͦC for 5min. To evaluate the PCR-amplified fragments, electrophoresis on 1.5% agarose gel was performed. Absence of PCR product for GSTM1 or GSTT1 was considered as null genotype and presence as positive genotype. The exon 7 of the constitutional CYP1A1 gene used as an internal control to confirm the null genotype and to avoid false negative reading.20
GSTM3 polymorphism: The PCR conditions were as follows: initial denaturation at 94 ͦC for 5min, 35 cycles of denaturation at 94 ͦC for 1 min, annealing at 55 ͦC for 1min, extension at 72 ͦC for 1min, and a final extension at 72 ͦC for 5min, using GSTM3 specific primers. After amplification of PCR products were subjected to restriction digestion by MnlI (37 ͦC, 18h) and resolved in 15% (w/v) polyacrylamide (100V, 2h), stained with ethidium bromide (0.5,µg/ml). DNA from GSTM3 A homozygote’s produced 11, 51, 86 and 125bp fragments (wild type pattern). PCR products from individuals with (GSTM3 A/GSTM3 B) produced 11, 51, 86, 125 and 134bp fragment, whereas MnlI digestions of GSTM3B/GSTM3B had revealed band pattern of 11, 125 and 134bp.8
GSTP1 polymorphism: PCR was carried out under following conditions: initial denaturation at 95 ͦC for 5 min, followed by 30 cycles of denaturation at 94 ͦC for 30s, primer annealing at 55 ͦC for 30s, extension at 72 ͦC for 30s. A final extension step of 72 ͦC for 5min was carried out to complete the elongation processes. The digestion of amplified product (176bp) with Alw261 (37 ͦC, 18h) produced 176bp (wild type), 85bp and 91bp (homozygous) and 176bp, 85bp and 91bp (heterozygous mutant). The digested products were separated in 15% (w/v) polyacrylamide gel stained with ethidium bromide.21
Statistical analysis
Comparison of patient and control allele frequency was done by x2 test and the Fisher exact test. An independent t- test was used to compare the GST level in the patients and controls. Data were analyzed with the clinical finding like age at onset, age at presentation, duration, sex, history of disease, facial weakness, dysphasia, dyspepsia, respiratory insufficiency, learning and writing disability (LWD), speech and languages disability (SLD), BMI, cataract, diabetes and SCK (serum creatine kinase), CK-MB (creatine kinase, isoform) and CK-MM (Creatine kinase, muscle isoform) using Pearson or Spearman correlation test. The statistical analysis was done using SPSS 16 version software and Graph Pad prism 5; p-values of less than 0.05 were considered significant.
The patient subjects fulfilled the clinical and molecular diagnostic criteria of DM1 (Figure S1) (Figure S2). The 27 DM1 patients with median age of 32.8years and their illness, family history and clinical features like facial weakness, dysphasia, dyspepsia, respiratory insufficiency, learning and writing skills, speech and languages skills, diabetes, SCK, CK-MB, CK-MM levels are shown in Table 1.
Variables |
GST activity r (p-value) |
Polymorphism |
|||
|---|---|---|---|---|---|
GSTM1 r (p-value) |
GSTT1r (p-value) |
GSTM3r (p-value) |
GSTP1r (p-value) |
||
Age at onset |
0.165(0.440) |
0.373(0.072) |
0.401(0.052) |
0.272(0.199) |
0.096(0.657) |
Age at presentation |
0.238(0.264) |
0.281(0.183) |
0.462*(0.023) |
0.011(0.961) |
0.021(0.922) |
Duration of disease |
0.043(0.843) |
-0.257(0.226) |
-0.062(0.773) |
-0.012177 |
-0.134(0.534) |
Sex |
0.134(0.534) |
-0.017(0.936) |
0.397(0.055) |
-0.122(0.569) |
0.263(0.214) |
History of disease |
-0.170(0.428) |
-0.192(0.370) |
0.228(0.285) |
0.035(0.872) |
0.116(0.588) |
Facial weakness |
0.228(0.284) |
0.247(0.245) |
0.162(0.451) |
-0.325(0.121) |
0.107(0.619) |
Dysphasia |
0.249(0.242) |
-0.076(0.726) |
-0.346(0.097) |
0.205(0.337) |
0.046(0.831) |
Dyspepsia |
0.111(0.604) |
-0.008604 |
0.000(1.000) |
0.130(0.546) |
-0.145(0.499) |
RI |
-0.207(0.332) |
-0.387(0.061) |
-0.308(0.144) |
-0.008(0.969) |
0.348(0.096) |
LWD |
-0.088(0.682) |
0.071(0.743) |
0.367(0.078) |
-0.146(0.497) |
0.266(0.209) |
SLD |
-0.211(0.322) |
-0.029(0.895) |
0.480*(0.018) |
0.170(0.426) |
0.225(0.289) |
BMI |
-0.140(0.513) |
0.031(0.884) |
0.309(0.141) |
-0.142(0.509) |
-0.212(0.320) |
Diabetes |
-0.006072 |
-0.098(0.650) |
-0.248(0.242) |
0.053(0.806) |
0.178(0.406) |
SCK |
-0.199(0.350) |
0.245(0.248) |
-0.256(0.227) |
0.333(0.112) |
-1.00(0.643) |
CK-MB |
-0.155(0.470) |
0.122(0.570) |
-0.376(0.070) |
0.397(0.055) |
0.287(0.174) |
CK-MM |
-0.206(0.333) |
-0.125(0.562) |
-0.266(0.209) |
0.327(0.119) |
-0.399(0.054) |
Table 1 Frequency of combination genotypes of GSTM1, GSTT1, GSTM3 and GSTP1 in DM1 patients and control groups.
*Correlation is significant at the 0.05 level (2-tailed).
GSTM1 and GSTT1 polymorphism and their correlation with clinical and laboratory findings
Genotype frequencies of GSTT1 and GSTM1 are summarized in Table 2 (Gel pattern figure not shown, Figure S3(A)&(B). DM1 Patients had significantly higher GSTM1*0 (GSTM1 null genotype) and GSTT1*0 frequency than controls (10/27 (37%) versus 8/100 (8%); p=0.0004); and (18/27 (66.67%) versus 8/100 (8%); p <0.0001). Deletion of the GSTM1 gene (in reference to presence of the gene) and GSTT1 were not associated with higher risk (OR 0.12; 95% CI=0.038-0.389 and OR 0.05; 95% CI=0.016-0.169) of DM1 than control (Table 2) (Figure S3(A)) (Figure S3(B)). The GSTM1 genotype had significantly negative correlated with dyspepsia (r= -0.48, p=0.018) while GSTT1 genotype positively correlated with age at presentation (r=0.46, p=0.023) and SLD (Table 1).
Polymorphism |
Genotype & group selected |
Present |
Null |
p-value |
OR (95% CI) |
|---|---|---|---|---|---|
GSTM1 |
DM1 Patients (n=27) |
17 |
10 |
|
0.1217 |
Control (n=100) |
92 |
8 |
0.0004 |
(0.038-0.389) |
|
GSTT1 |
DM1 Patients (n=27) |
9 |
18 |
|
0.052 |
Control (n=100) |
92 |
8 |
<0.0001 |
(0.016-0.169) |
|
Wild |
Hetero |
|
|||
GSTM3 |
DM1 Patients (n=27) |
8 |
19 |
0.1161 |
2.676 |
Control (n=100) |
14 |
86 |
(0.887-8.072) |
||
GSTP1 |
DM1 Patients (n=27) |
6 |
21 |
0.4405 |
0.595 |
Control (n=100) |
31 |
69 |
(0.198-1.789) |
||
Table 2 Frequency of GSTM1, GSTT1, GSTM3 and GSTP1 genotypes in DM1 Patients and Control groups
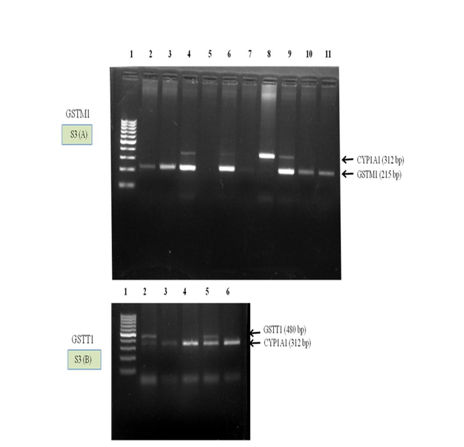
Figure S3 Gel patterns of genotyping of GSTM1 and GSTT1 genes.
(A) Polymerase chain reaction (PCR) result of GSTM1 gene with CYP1A1 as internal control. Lane 1 molecular weight marker (100 bp), Lane 5 null and Lane 2-4 and 6-11 GSTM1 genotype (wild type).
(B) PCR result of GSTT1 gene with CYP1A1 as internal control. Lane 1 molecular weight marker (100 bp), Lane 3, 4 and 6 null and lane 2 and 5 GSTT1 genotype (wild type).
GSTM3 and GSTP1 genotype, allelic frequencies and its correlation with studied parameters
The allelic frequencies of the GSTM3 and GSTP1 were in Hardy-Weinberg equilibrium in patients with DM1 and in controls (Gel patterns pics not shown, Figure S4 (A) & (B).
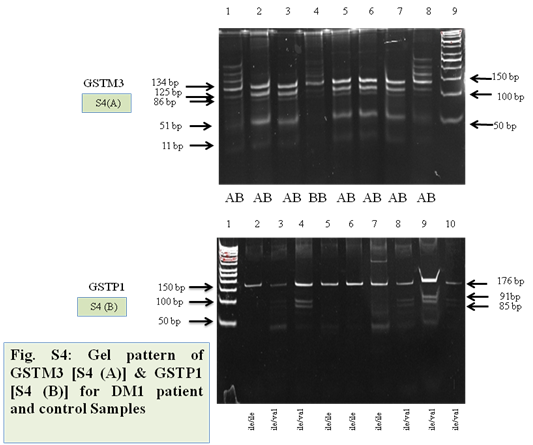
Figure S4 Representative gel images of GSTM3 and GSTP1 polymorphism.
(A) Lane 9 showing 50bp ladder, lane (1,2,3,5,6,7,8 and 9) heterozygous mutant and lane (4) wild type.
(B) Lane 1 showing 50bp ladder, lane (3,4,8,9 and 10) heterozygous mutant and lane (2,5,6 and 7) wild type.
DM1 patients showed lower genotype frequency of heterozygous GSTM3 (A/B) than control (19/27 (70.37%) versus 86/100 (86%); p=0.12) (Table 2 & Table 3). Mutation in the GSTM3 gene was associated with approximate 3-fold higher risk than control (OR 2.68; 95% CI = 0.89-8.07) (Table 2). The allelic frequencies were 0.46, 0.54 and 0.48, 0.52 for GSTM3*A and GSTM3*B in case of DM1 patients and control group respectively (Table 2). GSTM3 was significantly negative correlate only with duration of disease (r=-0.45, p= 0.027) (Table 1). The distribution of GSTP1 genotype and allelic frequencies are summarized in Table 2. The allele and genotype frequencies were comparable among patients and controls. The presence of two GSTP1 alleles (isoleucine and valine) confirmed that the wild-type pattern comprised individuals homozygous for either the GSTP1A (ile105/ile105) or GSTP1B (val105/val105) while the atypical pattern comprised heterozygotes for GSTP1A/GSTP1B (ile105/val105) alleles. Similar to GSTM1 and GSTT1, DM1 Patients had higher GSTP1 heterozygous frequency (ile/val) than controls (21/27 (77.78%) versus 69/100 (69%); p=0.44) and was not associated with risk (OR 0.59; 95% CI=0.19-1.79) (Table 2). The allelic frequencies were 0.61, 0.39 and 0.55, 0.45 for ile (A) and val (G) in case of DM1 patients and control group respectively (Table 3). Moreover, GSTP1 was not associated with any of the studied parameter (Table 1).
Gene |
Genotype |
Allele |
DM1 (n=27) |
Control (n=100) |
|---|---|---|---|---|
GSTP1 |
AA or ile/ile |
|
6 (22.22%) |
20 (20%) |
AG or ile/val |
|
21 (77.78%) |
69 (69%) |
|
GG or val/val |
|
Zero (0%) |
11 (11%) |
|
|
ile |
0.61 |
0.55 |
|
|
val |
0.39 |
0.45 |
|
GSTM3 |
AA |
|
3 (11.11%) |
5 (5%) |
AB |
|
19 (70.37%) |
86 (86%) |
|
BB |
|
5 (18.52%) |
9 (9%) |
|
|
A |
0.46 |
0.48 |
|
|
B |
0.54 |
0.52 |
Table 3 GSTP1 and GSTM3 genotypes and allele frequencies in DM1 patients and control groups
Genotypes |
Groups |
|
|---|---|---|
DM1 (n=27)* |
Control (n=100)^ |
|
+/+/+ (%) |
2(10%) |
10(13.33%) |
-/-/-/ (%) |
4(20%) |
Zero (0%) |
+/-/- (%) |
6(30%) |
6(8%) |
-/+/- (%) |
1(5%) |
5(6.67%) |
-/-/+ (%) |
2(10%) |
Zero (0%) |
+/+/- (%) |
3(15%) |
54(72%) |
+/-/+ (%) |
1(5%) |
Zero (0%) |
-/+/+ (%) |
1(5%) |
1(1.330%) |
-/-/-&ile/val (%) |
3(15%) |
Zero (0%) |
+/-/-&ile/val (%) |
5(25%) |
6(8%) |
-/+/-&ile/val (%) |
1(5%) |
3(4%) |
-/-/+&ile/val (%) |
2(10%) |
Zero (0%) |
+/-/-&ile/ile (%) |
1(5%) |
Zero (0%) |
-/+/-&ile/ile (%) |
1(5%) |
2(2.670%) |
-/-/+&ile/ile (%) |
2(10%) |
Zero (0%) |
Table 4 Abbreviations: +/+/+: GSTM1, GST1 and GSTM3 present; -/-/-: GSTM1, GSTT1 and GSTM3 absent; ile/val and ile/ile = heterozygous and homozygous status of GSTP1.
-/-/- in reference to +/+/+: * versus ^ (p <0.01; OR 0.03, 95% CI 0.001-0.067), -/-/-&ile/val in reference to +/+/+: * versus ^ (p = 0.02, OR 0.03, 95% CI 0.001-0.894)GSTM1, GSTT1, GSTM3 and GSTP1 combination genotype frequencies
GSTM1, GSTT1, GSTM3 and GSTP1 different combination genotype frequencies are presented in Table 4 in fifteen possible combinations. The +/+/+ combination represented the presence of GSTM1, GST1 and GSTM3 genotype while the -/-/- combination represented absence of combination of GSTM1, GSTT1 and GSTM3. The ile/val (AG) and ile/ile (AA) correspond to heterozygous and homozygous status of GSTP1 along with different other combination of GSTM1, GSTT1 and GSTM3. The frequency of -/-/- and -/-/-& ile/val (both in reference of +/+/+) were significantly higher in DM1 patients than controls (4 (20%) versus zero (0%); p <0.01) and (3 (15%) versus zero (0%); p=0.02) but were not associated with risk (OR 0.03, 95% CI 0.001-0.067) and (OR 0.03, 95% CI 0.001-0.894) respectively (Table 4).
GST level and its correlation with clinical and laboratory findings
The serum GST level (83.43±5.79 Vs 104.15±14.47 nmol/ml/min, p<0.001) was significantly reduced in the DM1 patient group compared to the control group (Figure S5). The GST level had significantly negative correlated only with diabetes (r=-0.47, p=0.037) and not with other studied parameter (Table 1).
Correlation between GST activity and Gsts (GSTM1, GSTT1, GSTM3 and GSTP1) polymorphism
There was no correlation between GST activity and GSTs (GSTM1, GSTT1, GSTM3 and GSTP1) polymorphism (Table S2).
|
|
Polymorphism |
GST Activity |
|||
|---|---|---|---|---|---|---|
GSTM1 |
GSTT1 |
GSTM3 |
GSTP1 |
|||
GSTM1 Polymorphism |
Pearson Correlation |
1 |
0.131 |
-0.201 |
0.225 |
0.222 |
Sig. (2-tailed) |
|
0.542 |
0.345 |
0.289 |
0.296 |
|
N |
27 |
27 |
27 |
27 |
27 |
|
GSTT1 Polymorphism |
Pearson Correlation |
0.131 |
1 |
0.071 |
0.026 |
0.15 |
Sig. (2-tailed) |
0.542 |
|
0.742 |
0.902 |
0.484 |
|
N |
27 |
27 |
27 |
27 |
27 |
|
GSTM3 Polymorphism |
Pearson Correlation |
-0.201 |
0.071 |
1 |
-0.103 |
-0.057 |
Sig. (2-tailed) |
0.345 |
0.742 |
|
0.63 |
0.791 |
|
N |
27 |
27 |
27 |
27 |
27 |
|
GSTP1 Polymorphism |
Pearson Correlation |
0.225 |
0.026 |
-0.103 |
1 |
0.041 |
Sig. (2-tailed) |
0.289 |
0.902 |
0.63 |
|
0.849 |
|
N |
27 |
27 |
27 |
27 |
27 |
|
GST Activity |
Pearson Correlation |
0.222 |
0.15 |
-0.057 |
0.041 |
1 |
Sig. (2-tailed) |
0.296 |
0.484 |
0.791 |
0.849 |
|
|
N |
27 |
27 |
27 |
27 |
27 |
|
Table S2 Correlation between GST activity and GSTs (GSTM1, GSTT1, GSTM3 and GSTP1) polymorphism
The glutathione S-transferase (GST) super family of enzymes has a vital role in phase II of biotransformation of xenobiotics and in protection of cells from reactive oxygen species (ROS) by its ability to utilize substrates of a wide range of products of oxidative stress.7 The aim of the present study was to evaluate the role of GSTM1, GSTT1, GSTM3 and GSTP1 genes in DM1 patients and controls. To the best of our knowledge, this is the first report from India evaluating the role of polymorphisms of these genes in DM1 and is also the first report on GSTM1, GSTT1, GSTM3 and GSTP1 polymorphisms and their correlation with GST activity. In the present study, serum GST significantly reduced in the patient group compared to the control group. The decreased level of antioxidant enzyme GST involves in higher oxygen free radical production and thus cell undergoes stress condition i.e. normal functioning of the cell become hindered (disturbed homeostasis).22,23 The serum GST significantly correlated with diabetes only. Indigestion problem, learning, speech, and writing disability are often observed and diabetes mellitus occur in 5% of DM1 cases (http://www.faqs.org/health/Sick-V3/Muscular-Dystrophy-Symptoms and ibmmyositis.com).
GSTM1*0, GSTT1*0, GSTM3 (A/B) and GSTP1 ile/val polymorphisms are associated with lack of or reduced GST enzyme activity. Thus, individuals with these polymorphisms may have inefficient detoxification of several metabolites, ROS/free radicals, aromatic hydrocarbons, and to protect tissues against DNA damage7,9,24,25 and therefore developing the symptoms of DM1. DM1 Patients had significantly higher GSTM1*0 (GSTM1 null genotype) and GSTT1*0 frequency than controls and the frequency of deletion of the GSTM1 gene and GSTT1 were higher in comparison to control but not associated with higher risk. Individuals having homozygous deletions of GSTM1 or GSTT1 lack GSTs and therefore may be unable to detoxify the substances efficiently, which may increase the risk of somatic mutations.24,25 Overall, based on several reports, theses genotype increased the risk of several types of cancers.26,27 However, this is not a general rule because a protective effect of the GSTT1 null genotype has been previously described.26 The GSTM1 genotype had significantly correlated with dyspepsia while GSTT1 genotype positively correlated with age at presentation only.
This is the first reported characterization of allelic variation of GSTM3 in DM1. All samples analysed showed the presence of amplified DNA, indicating that GSTM3 does not demonstrate a null allele as found in GSTM1 and GSTT1.28‒30 DM1 patients showed lower genotype frequency of heterozygous GSTM3 (A/B) than control and this was not significantly associated with the pathophysiology of the disease. Our findings are similar with the previous finding on hepatocellular carcinoma31 and lung cancer.32 The allelic frequencies were 0.45, 0.55 and 0.47, 0.53 for GSTM3*A and GSTM3*B in case of DM1 patients and control group respectively. Mutation in the GSTM3 gene was associated with approximate 3-fold higher risk than control. The allelic and genotype frequency of GSTP1 was comparable between patients and controls. Although Patients had higher GSTP1 heterozygous frequency (ile/val) than controls yet was not statistically significant and associated with risk as well as any of the studied parameter. The defensive role of GSTP1 in present findings was similar with the previous studies on asthma.33‒35
There was no correlation observed between GST enzyme activity and the studied GSTs polymorphism. However, to the best of our knowledge there is no such data and this aspect needs further investigation. The group of combination genotype frequency had no impact on risk of disease. The key finding of the present study is that the GSTM1 and GSTT1 active genotype is significantly associated with DM1. The genetic polymorphisms of GSTs genes differ significantly among racial groups and residential populations in different parts of the world.36,37 Some limitations of the study need to be drawn. A relatively small population was recruited and this finding needs to be replicated in a larger sample. Further research should also examine whether specific phenotypic characteristics, such as symptom profile, and treatment response are associated with the GSTs polymorphism.
In conclusion, our finding reported a significant association between DM1 and GSTT1 & GSTM1 polymorphism, suggesting that GSTT1 and GSTM1 seem to be a candidate gene for susceptibility to DM1 in Indian population. Further studies using genetic polymorphisms of glutathione or other antioxidant enzymes are required to clarify the relationship between increased oxidative stress and DM1. Nevertheless, large scales, more rigorous designs, especially studies stratified for gene–gene and gene–environment interactions on these polymorphisms and DM1 risk are needed to research, which may eventually lead to better comprehensive understanding of their possible roles in DM1.
Authors are thankful to Sanjay Gandhi Post Graduate institute of Medical Sciences, Lucknow and Department of Biotechnology, New Delhi, India for providing infrastructure facility to carry out laboratory work. Ashok Kumar is DBT-fellow did analysis of all the samples and Prof Sarita Agarwal has conceived the idea and provided financial support for acquisition of the task. Prof. Sunil Pradhan providing clinical support.
The authors are thankful to Sanjay Gandhi Post Graduate institute of Medical Sciences, Lucknow for providing infrastructure facility. Ashok Kumar is thankful to DBT-New Delhi (DBT-JRF 2009-10/515) for his fellowship.
Author declares that there is no conflict of interest.

©2015 Kumar, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.