Journal of
eISSN: 2373-4469


Book Review Volume 5 Issue 1
Department of Medicine, Imperial College London, UK
Correspondence: Kaoutar Bentayebi,Phage Therapy Group, Division of Brain Sciences, Department of Medicine, Imperial College London, Hammersmith Hospital Campus, London W12 0NN, UK, Tel +07388288672
Received: June 16, 2017 | Published: September 7, 2018
Citation: Bentayebi K, Hajitou A. A revised root for the human Y chromosome differentiation and diversity landscape among North African populations. J Investig Genomics. 2018;5(5):35-37. DOI: 10.15406/jig.2018.05.00075
North Africa is a crucial area in the settlement history of modern humans because it represents a possible connection between Africa and Europe. So far, genetic data were inconclusive about the fact that this region constitutes a barrier to gene flow, as previous results were highly variable depending on the genetic locus studied. The present study evaluates the impact of North Africa in reducing gene flow between populations from Africa and Europe, by comparing formally various genetic loci in the Y chromosome and analyzing several parameters of population differentiation taking into account the effects of both demography and natural selection at some loci. The Y chromosome is inherited through the paternal line and remains virtually unchanged through many generations. The combination of the analysis of Y chromosome STR and SNP will allow the analysis of human evolution in paternal lineages in different time scales. The forensic implication of the results are discussed.
Keywords: North Africa, Y chromosome, STR, SNP, ancestry, population, gene flow
The Y chromosome has a curious role in population Genetics and forensic genetics. It is male specific and constitutively haploid. It passes from father to son, and, unlike other chromosomes, largely escapes meiotic recombination. Two segments (the pseudoautosomal regions) do recombine with the X, but this amount to less than 3 Mb of its ~60-Mb length, which makes it easier to study than the autosomes.1 These traits, explains its use in an increasing number of studies, especially those that address the history of the human population and makes it easier to study than the autosomes. Nevertheless, the Y chromosome continues to be overshadowed in these studies by the autosomal chromosome and mtDNA, despite containing far more genetic information than either. One reason for the lagging position of the Y chromosome has been a delay in the generation of useful data. The first PHYLOGENETIC TREE for human mtDNA was published in 1987,2 and the first for the Y chromosome was published in 1989.3 Since then, Y-chromosome-based studies have slowly accumulated; with the maturation of sequencing and genotyping technologies, a tremendous increase in all kinds of genetic data looms. It is therefore an appropriate time to reflect on what the Y chromosome can offer to population-genetic studies.
The western Mediterranean region has a wide genetic interest in population genetics and forensic genetic studies. It has been a focus of genetic research for many years, due to its geostrategic position distinguished by its ancient and shifting history. The sequence differentiation of human Y chromosomes has occurred during and after the process of colonization and diffusion into the different geographic regions and continents, and its dissection is a useful tool for the investigation of range expansions, migrations and other forms of gene flow in prehistoric and historic times. In other words, the sequence variation of modern Y chromosomes represents a unique record of the history of human males, and of the relationships between past populations. A number of recent studies carried out in the Moroccan, Algerian and Tunisian populations have shown that most Y chromosomes can be classified into monophyletic units (haplogroups), which tend to be specific to each continent and major ethnic group.4–6 This study takes advantage of this ethnic/geographic specificity to define better the origins and relationships of the populations living in the Western Mediterranean basin.
We are interested in using two types of YDNA markers in our study: STR and SNP. Applications of these markers include phylogeny, forensics, paternity tests, genetic marker for disease, gene mapping, selection of traits in breeding, etc. i) STR stands for "short tandem repeat". The STRs are repetitions of 2 or more nucleotides that occur mostly in the non-coding parts of the DNA. The number of repeats is called allele value. STRs are highly polymorphic which means that the amount of repetitions in a specific STR can be different from person to person. The same number of repeats between persons across a wide range of STR markers indicates that they are most likely related. ii) SNP stands for "single nucleotide polymorphism". This means that there is a variation or mutation in one single nucleotide at a specific location in the genome (either in a gene or in a non-coding region). Unlike Y-STR markers which can have multiple repeats, SNP markers often only exist in 2 forms – the ancestral and the mutated allele. Y chromosome-specific single nucleotide polymorphisms (SNPs) are particularly useful for identification of stable paternal lineages because of their low rate of parallel and back mutations. By testing highly polymorphic Y-STR markers that can inform of recent ancestry in addition to the Y-SNP markers an even more detailed DNA profile can be obtained.7–9 Data here were collected from previous analysis on three North African populations (Morocco, Algeria and Tunisia). Y chromosomes were typed using 17 Short Tandem Repeats polymorphisms and SNP polymorphism for 52 Tunisian,10 102 Algerian11 and 267 Moroccan.12,13 All 421 individuals were tested for seventeen STRs (DYS19, DYS385 a/b, DYS389 I/II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438, DYS439, DYS448, DYS456, DYS458, DYS635 (Y GATA C4), and YGATA H4. dinucleotide repeat markers (YCAIIa, YCAIIb, DYS413a, and DYS413b).
From the extreme north western Africa to the east, data showed a decreasing pattern of diversity. The Moroccan, Algerian and Tunisian populations showed the respective values of different haplotypes: 257 (96.25%), 93 (91.18%) and 39 (75%) of which 247, 88 and 29 were unique. Forensic parameters displayed in table 1 showed a striking difference in discrimination capacity among the three North African populations for the 17 Y-STR markers, with a low value in Tunisia (0,750) compared to Algeria (0.918) and Morocco (0.963). Generally, lower levels or variation are observed on the Y chromosome in the Tunisian population. The smaller sample size of the Tunisian population included in this study could have affected the results and lead to show a lower diversity within this population. We couldn’t increase de sample size because we didn’t find studies in the literature describing wider Tunisian population sample. Further analysis on this population including these markers has to be done. The increasing pattern of genetic diversity from the North eastern part of Maghreb to the western shores, may be seemingly be explained by the out of Africa model. Indeed, the genetic segments of Y chromosome mighthad further time interval to acquire genetic differences and variation in Morocco compared to Tunisia. Two microvariant alleles (18.2 and 19.2) were observed at the locus DYS458 in the three studied populations, with a lower frequency than that observed in the Upper (Southern) Egyptian population. Although there are data suggesting that locus DYS458 may be affected by a higher mutation rate compared to other tetrameric Y-STR loci,14 the observed microvariants seem to reflect a single mutational event (Table 1).
Population |
Sample |
UH |
DH |
GD |
HD |
DC |
Morocco |
267 |
247 |
257 |
DYS385: |
0.9991 |
0.963 |
Algeria |
102 |
88 |
93 |
DYS458: |
0.997 |
0.918 |
Tunisia |
52 |
29 |
39 |
- |
- |
0.750 |
Table 1 Statistical parameters of 17 Y-chromosomal short tandem repeat loci in 421 North African male samples
*Abbreviations: UH, unique haplotype; DH, different haplotype; GD, gene diversity; HD, haplotype diversity; DC, discrimination capacity.
In order to get a deeper insight on the Y chromosome diversity in the Maghreb region, 41 Y-SNP were analyzed in a Tunisian population, and 22 biallelic markers respectively in Algeria and Morocco. The haplogroup E-M81, formally, E1b1b1b, E3b1b and E3b2 was the major Y chromosome haplogroup abundantly and respectively found in Morocco, Tunisia and Algeria. This haplogroupis the most common Y chromosome haplogroup in the Maghreb, dominated by its sub-clade E-M183. It is thought to have originated in the area of North Africa 5,600 years ago.15,16 This haplogroup reaches a mean frequency of 42% in North Africa, decreasing in frequency from approximately 80% or more in some Moroccan Berber populations, including Saharaw is, to approximately 10% to the east of this range in Egypt.16–18 Because of its prevalence among these groups and also others such as Mozabite, Middle Atlas, Kabyle and other Berber groups, it is sometimes referred to as a genetic "Berber marker". Pereira et al.,19 report high levels amongst Tuareg in two Saharan populations - 77.8% near Gorom-Gorom, in Burkina Faso, and 81.8% from Gosi in Mali. There was a much lower frequency of 11.1% in the vicinity of Tanut in the Republic of Niger. E-M81 is also quite common among North African Arabic-speaking groups. It is generally found at frequencies around 45% in coastal cities of the Maghreb (Oran, Tunis, Tizi Ouzou, Algiers).11,16 It is also seen, although at low frequency, in the Iberian Peninsula (4%) and Sicily (3%) due to recent gene flow from Northwest Africa through Gibraltar strait.20,21 The second most abundant haplogroup in the studied population was J1 that showed a decreasing prevalence from Tunisia to Morocco with respectively, 35%, 22.5% and 10%. The J1 haplogroup, defined by the single nucleotide polymorphism (SNP) M267, is most frequent in the Arabian Peninsula especially in Yemen (76%).22 This could be attributed to the early medieval period during which the Semitic expansion spread J1 out of Arabia into North Africa (Figure 1).23
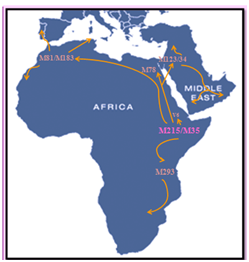
Figure 1 Distribution and movement of the major E-M215 lineage.
E-M215 also known as E1b1b was dated by Cruciani in 2007 to about 22,400 years ago in the Horn of Africa. E-M215 is defined as one of the major patriline ages of humanity, supporting the “out of Africa” theory. Recent discovery showed that the earliest homo sapiens was found in Morocco and has about 300.000 years old. In Africa, E-M215 is distributed in highest frequencies in the Horn of Africa and North Africa, whence it has in recent millennia expanded as far south as South Africa, and northwards into Western Asia and Europe (especially the Mediterranean and the Balkans). Studies on Y chromosome showed that EM81 is the most frequent lineage in North Africa. E-M78 is more present in the east and decline toward the west in contrast with E-M81.6,24–28
The analysis of a large panel of Y-STR and Y-SNP are very useful in population genetics to characterize the male lineage of human populations and constitute a benefic tool to forensic expert. The previous studies showed that the genetic background of Maghrebien populations is very similar and close each other. The genetic diversity has an increasing pattern from East to west of North Africa, seemingly supporting the hypothesis of out of Africa model. Most results showed that the Maghrib is characterized by the predominance of the EM-81 haplogroup that is specific and native to this region. Its presence in some region of southern Europe may be due to the colonization of North African Arabic and berbers to the Iberian Peninsula through the strait of Gibraltar. The forensic values encourage widely the use of these markers in forensic genetics to resolve some complex kinship cases. Also, haplotype can help the investigators to infer the provenience of the subject who left the evidence. At the same time, information on the geographic and ethnic distribution of Y-SNP haplogroups and their internal YSTR variability can assist the expert with the estimate of the frequency of a particular haplotype in populations for which a specific reference database of Y-STR haplotypes isnot yet available. This study confirmed that precious informations might come both from YSNPs haplogroup distribution and microsatellite variability to characterize the genetic background of the North West African population.
None.
The author declares that there is no conflict of interest.

©2018 Bentayebi, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.