Journal of
eISSN: 2373-4469


Research Article Volume 5 Issue 1
1Unita Operativa Complessa di Cardiologia-ASSL3 Nuoro ATS Sardegna, Italy
2Laboratorio Specialistico di Ematologia-ASSL3 Nuoro ATS Sardegna, Italy
3Universita del Molise Sede, Facolta di Medicina, Italy
Correspondence: Nicola Marziliano BD PhD,UOC Cardiologia-PO San Francesco, ASSL3 Nuoro-ATS Sardegna, Via Mannironi 1-08100 Nuoro, Italy, Tel +393495982624
Received: November 21, 2017 | Published: February 26, 2018
Citation: Marziliano N, Gaddeo C, Orrù V, et al. A heterozygous mutation of the elastin gene (eln:p.val154met) fully co-segregates with classic marfan phenotype in a sardinian family: a possible novel disease-gene?. J Investig Genomics. 2018;5(1): 00072. DOI: 10.15406/jig.2018.05.00072
Fibrillin 1 (FBN1), TGF-β-receptor 2 (TGFBR2) and Latent binding protein (LTBP) gene defects have been associated with Marfan syndrome (MFS) with classic phenotype. However, molecular testing in adults with overt MFS phenotype does not reach the 100% of genetic diagnosis.
We then designed a targeted NGS panel by pooling 20 causative genes involved in the Extra Cellular Matrix (ECM) constitution and remodelling with the aim of increase the molecular yield when testing MFS patients. We then run molecular analysis on a PGM Io Torrent platform and confirmed the identified sequence variants by conventional Sanger Sequencing on MFS diagnosed according to the Ghent criteria. We identified a heterozygous missense mutation in the Elastin (ELN) gene (p.V154M) in a Sardinian family with classic MFS phenotype. The co-segregation analysis of the identified missense mutation proved to be pathogenic within the family.
Our data, although on a single case/family, led us to conclude by proposing a novel association of the ELN gene mutations to contribution of the onset of MFS classic phenotype.
Keywords: marfan syndrome mfs, elastin ELN gene, next generation sequencing
FBN1, fibrillin 1; TGFBR2, TGF-β-receptor 2; LTBP, latent binding protein; ECM, extra cellular matrix; MRI, magnetic resonance imaging
Marfan syndrome (MFS, MIM #154700) is an autosomal dominant disorder of the extra cellular matrix affecting the connective tissue with cardiovascular, skeletal, neurological, integumental and ocular abnormalities.1 More than 85% of MFS cases are causally linked to mutations in the fibrillin-1 gene (FBN1, 15q21.1). In 1994, a second locus (MFS2) (MIM #154705) was found to map at 3p25–p24.22 after the identification of a family with autosomal dominant MFS not linked to FBN1 gene harbouring a 3p24.1 chromosomal breakpoint disrupting the transforming growth factor beta type II receptor (TGFBR2) gene in a Japanese individual with MFS. Subsequently additionally reports showed association of TGFBR2 mutations with MFS.2−4 Presently 3 genes (FBN1, TGFBR2 and LTBP) are associated with MFS classic phenotype presenting different degrees of clinical signs expression and variability (Table 1).4,5
Disorders |
Genes |
Features |
MFS |
FBN1, TGFBR2, LTBP |
Skeletal, ocular and cardiovascular defects. Aortic dilation occurs at the level of sinuses of Valsalva |
LDS |
TGFBR1, TGFBR2, SMAD3, TGFB2, TGFB3 |
LDS all forms. Aggressive TAA |
vEDS |
COL3A1 |
Risk of spontaneous intestinal, uterine and arterial rupture as well as joint and cutaneous manifestations. |
TAAD |
ACTA2, MYH11, TGFB2, MYLK, PRKG1 |
Livedo reticularis, iris flocculi, patent ductus arteriosus, nonthoracic aneurism, premature coronary artery disease, ischemic strokes and moyamoya |
Table 1 List of the diseases, their genetic background and their principal clinical features. MFS, marfan syndrome classic phenotype; LDS, loeys-diets syndrome; TAA, thoracic aortic aneurysm; vEDS, vascular ehlers-danlos syndrome; TAAD, thoracic aortic aneurysm and dissection
MFS and other collagen-disorders (i.e. vascular Ehlers-Danlos syndrome or vEDS, Loeys-Diets syndrome(s) or LDS) share histopathologic features of medial degeneration, characterized by destructive matrix remodelling with elastin fragmentation, impaired proliferation of vascular smooth muscle cells, and proteoglycan deposition.1 Because the genes mutated in MFS and vEDS encode for extra cellular matrix (ECM) proteins (fibrillin-1 and type III procollagen, respectively) and other genetic conditions leading to aortopathy show causative mutations in small molecules involved in the ECM remodelling belonging to the TGF-b pathway (thoracic abdominal aortic aneurysm or TAA and abdominal aortic aneurysm or AAA), a plausible hypothesis is that altered ECM contents led to structural weakness in the aortic wall, thereby leading to progressive aneurysmal dilation (Figure 1). Although this assumption is too simplistic, genetic evidences supported this hypothesis with a plethora of additional mechanistic features (Figure 1)(Table 1).
We here report a novel elastin (ELN, 7q11.23) gene missense mutations (p.Val154Met) found in a Sardinian family presenting with familial autosomal dominant MFS characterized by major cardioskeletal signs, and in one young lady with skeletal and neurological involvement. In all of the family members the screening of known genes associated with aortopathies tested negative. We conclude by proposing a novel association of the ELN gene mutations to contribution of the MFS classic phenotype.6
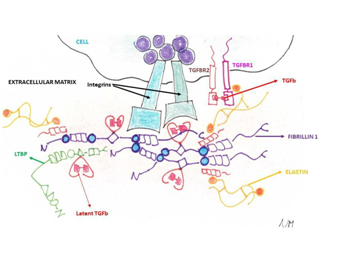
Figure 1 The ECM and the cellular surface components involved in the collagen disorders and their relationships. The chemokine Transforming Growth Factor-b (TGF-b) has a pivotal role being sandwiched in the fibrillin (FBN1 gene) and latent binding proteins (LTBP gene) and being the ligand of the Transforming Growth Factor-b Receptor 1 (TGFBR1 gene) and Transforming Growth Factor-b Receptor2 (TGFBR2 gene). Additionally, elastin fibres (ELN gene) are bound by S-S bridges to the fibrillin microfilaments.
The proband is a 46-year-old male (III:5; (Table 2)(Figure 2)), whose mother (II:3) is affected by MFS clinically diagnosed cording to the Ghent criteria7 with aortic dissection surgically corrected at the age of 40 years. The patient was first diagnosed with MFS at age 35 due to an ascending aortic dissection with a severe myopia (ectopia lentis). Since then, several cardiological clinical and echocardiographic evaluations were repeated. His phenotype is now characterized by aortic dissection, mitral valve prolapse with mild incompetence (regurgitation), major skeletal signs, including pes planus, arachnodactily with wrist and thumb signs, scoliosis and left lumbar gibbus, ocular (ectopia lentis) and nervous systems (dural ectasia) also show major signs. The Ghent criteria for diagnosis were fulfilled, independently on the family history. The molecular analysis by means of a Next Generation Sequencing with a targeted panel of genes (see Methods) identified the presence of a heterozygous missense mutation p.V154M in the elastin gene (ELN;NM_001278939.1 c.2955C>A p.Val154Met in Figure 3A; coverage 72X). The same mutation was validated by Sanger sequencing and then found in the proband’s family members presenting with a variability of clinical signs of MFS (Table 2) showing an autosomal dominant inheritance with phenotypic variability. The frequency of the mutated allele as from the consortium ExAC (Exome Aggregation Consortium) was of 23 counts on 118634 alleles (allele frequency of 0.2%; http://exac.broadinstitute.org/dbsnp/rs145669576) thus supporting his possible pathogenetic rle as highlighted by provisional algorithms such as PolyPhen and SIFT (see methods). Furthermore, this sequence variant was absent in a consecutive series of about 300 patients genotyped in our centre and referred for collagen-related disorders.8
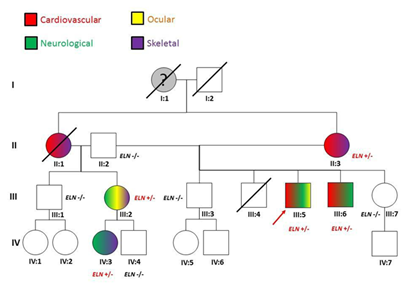
Figure 2 The proband’s (III:5) family pedigree. For each of the tested member, phenotypic (colour code) and genotypic information are given. The grand-mother (I:1) was reported died suddenly aged 55. No autpsy was done. II:1 died during in a car accident: she was reported presenting skeletal and cardiovascular signs (mitral valve prolapse and aortic dilation).
Systems and clinical features |
II:2 |
II:3 |
III:1 |
III:2 |
III:3 |
III:5 |
III:6 |
III:7 |
IV:3 |
IV:4 |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
Age (years) |
82 |
76 |
58 |
56 |
48 |
46 |
46 |
41 |
17 |
15 |
|
Sex |
M |
F |
M |
F |
M |
M |
M |
F |
F |
M |
|
Skeletal |
Arachnodactyly |
- |
- |
- |
+ |
- |
+ |
+ |
- |
- |
- |
Dolichostenomelia |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
Joint laxity |
- |
- |
- |
+ |
- |
- |
- |
- |
- |
- |
|
Scoliosis |
- |
+ |
- |
+ |
- |
+ |
- |
- |
+ |
- |
|
Pectus deformity |
- |
- |
- |
+ |
- |
- |
- |
- |
- |
- |
|
Limited elbow extension |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
Protrusio acetabulae |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
Facial dysmorphism |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
High-arched palate |
- |
+ |
- |
+ |
- |
- |
- |
- |
+ |
- |
|
Dental malocclusion |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
Pes planus |
- |
+ |
- |
+ |
- |
+ |
+ |
- |
+ |
- |
|
Orthopedic surgery |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
Ocular |
Ectopia lentis |
- |
- |
- |
+ |
- |
+ |
- |
- |
- |
- |
Myopia |
- |
+ |
- |
+ |
- |
- |
+ |
- |
+ |
- |
|
Cataract |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
Retinal detachment |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
Glaucoma |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
Cardiac |
Dilatation of the ascending aorta |
- |
+ |
- |
- |
- |
+ |
+ |
- |
- |
- |
Dissection of the ascending aorta |
- |
+ |
- |
- |
- |
+ |
- |
- |
- |
||
Dilatation or dissection of the descending or abdominal aorta before age 50 years |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
Mitral valve prolapse |
- |
+ |
- |
+ |
- |
- |
+ |
- |
- |
- |
|
Mitral regurgitation |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
Aortic insufficiency |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
Aortic surgery |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
Isolated valvular surgery |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
Skin |
Striae |
- |
- |
- |
+ |
- |
- |
- |
- |
+ |
- |
Herniae |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
Lung |
Pneumothorax |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
CNS |
Dural ectasia |
- |
- |
- |
+ |
- |
+ |
- |
- |
+ |
- |
Molecular genetics |
|
|
|
|
|
|
|
|
|
|
|
Table 2 The clinical characteristics of the proband (III:5; in grey) and his family members (see Figure 2 for the full pedigree). Clinical features are displayed according to the Ghent nosology7. Aortic dilation (> 45mm); CNS (Central Nervous System)
Mutations of the ELN gene (7q11.23) are presently associated to:
Our results, although on a single family, support the ELN gene as a candidate for FBN1 and TGFBR2-negative patients diagnosed with MFS according to Ghent's criteria.7
The clinical problem of differential diagnosis between MFS and MASS syndrome (myopia, mitral valve prolapse, mild aortic dilatation, and skin and skeletal manifestations (MIM #157700))12 in these patients is particularly difficult. We excluded a MASS syndrome in our proband because the skeletal system was severely affected along with the cardiovascular system and his aunt (II:1) deceased for aortic root dissection at 40 years of age. Additionally, in our proband the major cardiovascular, skeletal and ocular signs led per se to the diagnosis of MFS. Therefore since no other mutations of the presently known genes were identified and because of the co-segregation within the family, we might support a novel association of the ELN gene to MFS (analogously to several other diseases that are allelic at the same locus): the major clinical message is that aortic aneurysms and dissections, valvular stenosis and skeletal features are the common marker of all known phenotypes associated to ELN gene defects.
Available clinical and scientific data on this single family, however, are still nonsufficient to exclude the presence of deep intrinsic mutations, large genes re-arrangements in known genes as well as the presence of mutations in genes yet to be identified. Furthermore, additional studies on the involved extracellular matrix components by means of conventional histochemistry, immunohistochemistry and electron microscopy can shed lights on the intimal mechanisms by which ELN gene defects can underlie such phenotypes.
The patients
The clinical characteristics of the proband and his family members are summarised in Table 2. All patients were referred to our outpatient’s clinic for Inherited Cardiovascular and they underwent multidisciplinary clinical and non-invasive instrumental study (cardiological, ophthalmological, orthopedic, radiological, and lumbo-sacral magnetic resonance imaging (MRI) for the detection of dural ectasia). The aortic root diameter was evaluated with 2D echocardiography by experienced echocardiographists. The diagnostic criteria for MFS were still based on Ghent nosology in adults.7 Patients received detailed information on the MFS and related diagnostic work-up, likelihood of mutation identification, methods of analysis of the genes and related sensitivity. Written informed consent was obtained from all probands or, in case of age<18 years, from their parents, as well as from all relatives who underwent clinical and genetic screening.
Molecular genotyping
DNA isolation: Genomic DNA was isolated from peripheral blood leukocytes by means of the semi-automated platform Maxwell 16 (Promega).
Next generation sequencing: A custom panel was designed with the AmpliSeq chemistry (Thermofisher; design identification code IAAD#93109). The panel includes the complete coding regions (with exon/intron boundaries) of the following disease-genes: FBN1, TGFBR1, TGFBR2, TGFB2, TGFB3, MYH11, ACTA2, MYLK-AS1, MFAP5, PRKG1, SMAD2, SMAD4, COL1A1, COL1A2, COL5A1, COL5A2, COL3A1, COL5A3, LTBP and ELN for a total of 784 amplicons with a target gene coverage of 98.6%. The preparation of the amplification libraries was fully automated and run on an Ion Chef platform (Thermo fisher) and then the Ion 318 chips (version 2) were run on an Ion PGM (Thermo fisher). We then annotated rare (allele frequency<1%) damaging mutations included loss of function (i.e. nonsense, canonical splice site, and frameshift) and missense variants annotated as pathogenic or predicted to be damaging by computer algorithms and public databases (Anno Var, Sift, PolyPhen, Clin Var, HGMD, Variant Interpreter and Variant Caller). After mutations’ prioritisation by bio-informatics analysis, the candidate genes mutations were subsequently validated (see later).
Sanger sequencing and co-segregation analysis: To validate and perform co-segregation analysis, we amplified the exon 23 of the ELN gene with the following intron-specific primers design by the program Primer Express (Thermo Fisher) under stringent conditions: ELN23F5’TCGACTGCACCATTTTACAAATGG-3’ -and ELN23R 5’-CCACACTCCTAGAGGTCATTCATCA-3’. The same primers were applied to the PCR products for the Sanger sequencing done with the Big Dyes chemistry version 3.1 on a ABI PRISM 3500 platform (Thermo Fisher) (Figure 3). Sequence analysis and alignments were done by means of SeqA (version 6.0) and Seq Scape (version 3.5) respectively (Figure 3B).
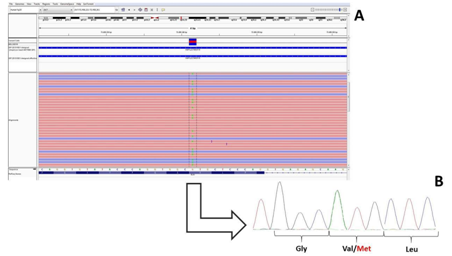
Figure 3 A) Next Generation Sequencing reads as from the IGV (integrative Genomic Viewer) software of the Exon 23 of the ELN gene (in the proband III:5) being G the wild type allele and A the mutate allele; B) Sanger Sequencing confirmation of the NGS data showing the double peak at the Val 154 amino acidic residue.
This report was supported by the Regione Sardegna project “Epidemiologia e Genetica della morte improvvisa in Sardegna e sue correlazioni con le canalopatie”.
There is no conflict of interest.

©2018 Marziliano, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.