Journal of
eISSN: 2373-4469


Case Report Volume 4 Issue 1
1Blacktown Clinical School and Research Centre, University of Western Sydney, Australia
2Clinical Pharmacology & Toxicology, Western Sydney Toxicology Service, Australia
3Discipline of Emergency Medicine, Sydney Medical School, Australia
Correspondence: Irina Piatkov, Blacktown Clinical School and Research Centre, Blacktown Hospital, Blacktown 2148, Australia
Received: October 31, 2016 | Published: January 4, 2017
Citation: Piatkov I, Mann G, Jones T, et al. Serotonin toxicity and cytochrome p450 poor metaboliser genotype patient case. J Investig Genomics. 2017;4(1):1-5 . DOI: 10.15406/jig.2017.04.00054
Serotonin toxicity commonly occurs in the context of serotonergic drug overdose or from interactions between multiple serotonergic agents. We describe a 29 year old woman with depression and anxiety who was commenced on regular fluoxetine 20mg/day for 7 weeks, followed by 40 mg/day for 3 weeks. During the 10-week period she began to have increasing agitation and episodes of flushing, sweating and tremor. She presented to the emergency department with a similar acute episode. Following cessation of fluoxetine and treatment with cyproheptadine and diazepam, her symptoms improved and she was discharged the following day. Informed consent was obtained from the patient for genetic testing of her CYP enzymes. Restriction Fragment Length Polymorphism assay (PCR-RFLP) revealed a presence of homozygote rs3892097 and rs1065852 polymorphisms in CYP2D6 and heterozygote 2C19 rs4244285/rs4986893 polymorphisms, which are associated with poor metaboliser phenotype and presence of one copy of CYP1A2 rs35694136 and rs762551. This patient genotype is a very rare case of combined loss-of-function of CYP2D6 and CYP2C19 isozymes. In addition, heterozygote polymorphisms, which are associated with impaired activity of CYP1A2, possibly contribute to low activity of the cytochrome P450 drug metabolising pathway. The involved cytochrome P450 isoforms exhibit genetic polymorphisms that affect their catalytic activity.
Keywords: Serotonin syndrome; Genetic polymorphism; Cytochrome P-450; CYP2D6; CYP2C19; CYP1A2
ED: Emergency Department; GP: General Practitioner; ST: Serotonin Toxicity; CYP: Cytochrome P450; SSRI: Selective Serotonin Reuptake Inhibitor
Serotonin toxicity developed by patient with loss-of function Cytochrome P450 genetic polymorphisms’ combination is described. Genotyping revealed CYP2D6, CYP2C19 and CYP1A2 non-functional polymorphisms previously associated with poor metaboliser phenotype.
Case
A 28-year-old female patient was admitted to the emergency department (ED) for anxiety and agitation including increased sweating of palms and restless legs. She had been treated with antidepressants by her general practitioner (GP) for 2 years. Her medication was switched 2 months previously and three weeks prior to presentation to ED, her GP increased the fluoxetine dose from fluoxetine 20 mg/day, to 40 mg daily. On admission, the patient was confused, irritable, restless and agitated. Her physical examination was significant for tachycardia (122 beats/minute), hypertension (systolic BP 147 mmHg) and agitation. Neurologic examination was positive for hyperreflexia and clonus. She denied thoughts of self-harm, was compliant with medications, alert and orientated. Her background included smoking 10-15 cigarettes per day, and she had a family history of anxiety (both her mother and brother); she had no known allergies. Routine laboratory investigations were normal except for hypokalaemia (K).
Serotonin toxicity was suspected and fluoxetine was ceased. Cyproheptadine 4 mg and diazepam 5 mg were administered to reduce agitation and serotonergic symptoms. Doxepin (50 mg daily) was recommended after 1 week medication-free time. Potassium replacement was given orally and the patient was discharged home on the following day.
This case represents a clinically diagnosed serotonin toxicity occurring during normal prescribed doses of fluoxetine. Poor metaboliser status was suspected and a blood sample was collected for the genotyping analysis with the patient’s consent. In addition, the Liquid Chromatography QTOF MS blood test revealed fluoxetine in concentration off 0.34 mg/L, however, norfluoxitine was not included in the MS test.
Serotonin toxicity
Serotonin toxicity (ST), or often referred to as serotonin syndrome, is a potentially lethal adverse effect due to serotonergic overactivity [1]. Often under-recognized, serotonin toxicity is an important diagnosis to make as the prognosis is favourable if detected early and complications are managed appropriately.
ST involves several serotonin receptor sub-types [1-6]. Potential mechanisms of serotonin toxicity include:
Based on the Clinical Information Access Portal data, specific agents that may be implicated in ST include: amphetamines and their derivatives, analgesics, antidepressants/mood stabilisers, monoamine oxidase inhibitors SSRIs, serotonin-norepinephrine reuptake inhibitors, serotonin 2A receptor blockers, St. John's Wort, tricyclic antidepressants, anti-emetics and anti-migraine drugs. Miscellaneous agents that may cause serotonin toxicity include cocaine, dextromethorphan, linezolid, l-tryptophan and 5-hydroxytryptophan [7].
In clinical practice, ST is diagnosed using the Hunter Serotonin Toxicity Criteria [8]. ST has most commonly been reported when patients concomitantly receive two or more serotonergic agents, for example a combination of a monoamine oxidase inhibitor and a monoamine reuptake inhibitor. Reports suggest that ST can occur with single agent overdose and even with therapeutic dosing [9-13]. Currently, no specific tests are available for the diagnosis of ST and blood serotonin levels do not correlate with clinical findings.
ST is a recognised adverse drug reaction to serotonergic psychotropic agents. As stated by Iqbal and co-authors [1], variations in drug response may be due to age, gender, morbidity, co-medication, food components, smoking and environmental factors. However, polymorphisms present in genes involved in drug metabolism, are responsible for most of the variations [14,15].
Several pharmacogenetic factors were identified by studies involved ‘pharmacokinetic pathways candidate gene approach’ and CYP enzymes, dopamine and serotonin gene variants have been linked with treatment-associated side effects of psychotropic drugs [14,15]. CYP 2D6, 2C19 and 2C9 are the most commonly studied cytochrome P450 enzymes. Based on the CYP nomenclature website per-reviewed data [16], the most frequent variations in Phase I metabolism of drugs are consequences of gene polymorphisms or duplications. Authors contributed it to the fact that nearly 80% of all drugs in current practice, along with most psychotropics, are metabolised via these pathways. According to previously analysed data, patients with drug-induced akathisia have a higher prevalence of abnormal metaboliser genotypes [17-19]. Serotonin pathway gene variants have also been linked to the development of adverse drug reactions, including the development of neurotoxicity [14,20-22].
Informed consent was obtained from the patient for genetic testing of her CYP enzymes. We have analysed variant alleles of CYP2D6*4 rs3892097, CYP2D6*5 (DEL), CYP2C19*2 rs4244285, CYP2C19*3 rs4986893, CYP2C9*2 rs1799853, CYP2C9*3 rs1057910, CYP1A2*1D rs35694136, CYP1A2*1F rs762551, CYP2D6*41 rs28371725, CYP3A4 rs2740574, CYP2D6*10(*4) rs1065852 that affect the function of cytochrome enzymes. DNA was extracted from blood using the manufacturer’s protocol for the QIAGEN EZ1 BioRobot system. The genotyping method involves specific restriction enzyme digestion of amplified PCR products (PCR-RFLP) and fragment analysis based on capillary electrophoresis Agilent Bioanalyser methodology described in our previous publication [23].
Restriction Fragment Length Polymorphism assay (PCR-RFLP) revealed a presence of homozygote rs3892097 and rs1065852 polymorphisms in CYP2D6 and heterozygote 2C19 rs4244285/rs4986893 polymorphisms, which are associated with poor metaboliser phenotype and presence of one copy of CYP1A2 rs35694136 and rs762551.
The constructive guidance for psychotropic drug prescription remains the subject of new developments in medicine. Nevertheless, the latest progress in science and biotechnology provides insights into drug pharmacokinetic and pharmacodynamic pathways. Drug metabolism can be analysed on the basis of genetic variations in metabolising enzymes and these variations can be tested in medical laboratories. Due to adverse drug reactions, which can sometimes even be life-threatening, prescription guidelines and recommendations are critical for drug safety in psychiatry.
Cytochrome P450
CYP is a group of oxidative/dealkylating enzymes, which are responsible for the primary metabolism of many drugs, accounting for about 75% of the total number of different metabolic reactions [24]. CYP enzymes are localised in the microsomes of many tissues, including the intestines and liver, and participate in hydroxylation or dealkylation of many commonly prescribed psychotropics such as antidepressants and antipsychotics. In principle, the majority of psychotropic drugs are metabolised by the cytochrome P450 (CYP) family of enzymes [25]. CYP-related metabolites of these drugs are highly chemically active molecules, so consequently they are likely to be associated with toxicity or unpredicted adverse reactions [26].
Genes for most CYP isozymes are extremely polymorphic, that is, they frequently contain altered DNA, which can produce variants of the enzyme, and when this occurs, the activity of the enzyme is affected. The genetic polymorphism in CYP enzymes is a major factor in the individual variability of drug metabolism [27,28].
Identified SNPs
rs3892097 (CYP2D6*4) is the most common non-functional allele of CYP2D6 gene. It was demonstrated that homozygote genotype results in a non-functional protein formation [29]. This non-functional allele was described in association with neuroleptic malignant syndrome [30], metoprolol pharmacokinetics/pharmacodynamics modulation [31], metoclopramide side effect [32], tamoxifen efficacy [33,34], colchicine efficacy [35], with environmental sensitivity-related illnesses [36], enhancing susceptibility to head and neck squamous cell carcinoma and chemotherapeutic response [37], increased incidence of systemic sclerosis [38] the risk of Parkinson’s disease [39] and Poor Metaboliser phenotype for several drugs [40]. Patients with the *4/*4 diplotype and depression may require a lower dose of citalopram, escitalopram, fluoxetine, fluvoxamine, paroxetine or sertraline as compared to patients with the *1/*1 genotype [40].
Rs1065852 variant is described in association with poor metabolisers of debrisoquine [29] amitriptyline, nortriptyline and fluvoxamine [41,42]. Thermal instabilities and reduced intrinsic clearance by the protein encoded by the rs1065852 (T) allele were described [43].
CYP2C19 rs4244285/rs4986893 combined effect on protein function create a decrease activity of cytochrome enzyme. It was demonstratedthat patients with two no function CYP2C19 alleles (*2/*3) may have decreased metabolism of mephenytoin [44], diazepam [45], esomeprazole [46]. Patients with the CYP2C19 *2/*3 genotype infected with Helicobacter pylori (H. pylori) may have an increased likelihood of eradication when treated with proton pump inhibitors [47].
It was demonstrated that CYP1A2 rs35694136 and rs762551 have a significant impact on clozapine and theophylline serum concentration [48,49].
Fluoxetine
According to the MIMS Online (Medicine Information portal) data, fluoxetine is a selective serotonin reuptake inhibitor (SSRI) and is used for the treatment of major depressive disorder, obsessive compulsive disorder, bulimia nervosa and panic disorder [50].
Fluoxetine is extensively metabolized in the liver by several cytochrome P450 enzymes with CYP2D6 being a major contributor [28]. At the same time, fluoxetine is an inhibitor of the CYP2D6 enzyme pathway and a potential dug-drug interaction substrate [51]. Co-administration of fluoxetine with other drugs that are metabolised by CYP2D6 can convert a normal CYP2D6 metaboliser to a poor metaboliser. Fluoxetine and norfluoxetine are inhibitors of CYP2D6 mediated reactions and demonstrated inhibitory potency toward CYP2C19, CYP2C9, and CYP3A4 in in vitro studies [52-54]. Several drugs metabolised by these enzymes, hence fluoxetine can influence metabolism and pharmacokinetics of coadministered drugs. Clinically relevant drug interactions have been reported with tricyclic antidepressants and neuroleptics through the interaction with isoenzyme CYP2D6 [55].
As recommended by the FDA label [56], “therapy with medications that are predominantly metabolized by the CYP2D6 system and that have a relatively narrow therapeutic index should be initiated at the low end of the dose range if a patient is receiving fluoxetine concurrently or has taken it in the previous 5 weeks. Thus, his/her dosing requirements resemble those of poor metabolizers. If fluoxetine is added to the treatment regimen of a patient already receiving a drug metabolized by CYP2D6, the need for decreased dose of the original medication should be considered”.
Fluoxetine's metabolism involves the Cytochrome P450 system (Figure 1), and a combination of drugs which are also metabolised by CYPs may lead to drug interactions, even if fluoxetine has been discontinued for 4-5 weeks [55,57-61].
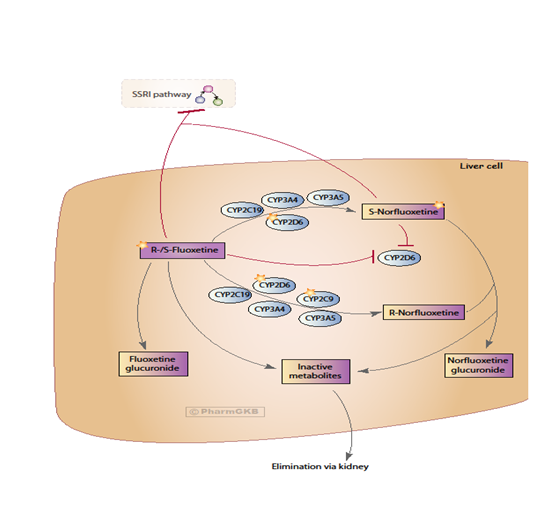
Fluoxetine is a racemic mixture of two enantiomers and its pharmacokinetics are complex. Norfluoxetine, an active metabolite of fluoxetine, is formed by demethylation [62]. S-fluoxetine is more potent in the inhibition of serotonin reuptake than R-fluoxetine. However, the active metabolite S-norfluoxetine has about 20 times higher reuptake blocking potency than its R-enantiomer [63]. S-fluoxetine, R-fluoxetine, S-norfluoxetine and R-norfluoxetine also have differential kinetics. The Fuller and co-authors demonstrated that that after several weeks of treatment, the plasma concentration of both S-enantiomers is about two times higher than the concentration of the R-enantiomers [62].
Fluoxetine is eliminated mostly through oxidative metabolism and conjugation [62] and excreted in urine with less than 10% excreted unchanged or as fluoxetine glucuronide [64]. Majority of the metabolic end products are unknown.
As described by some authors, CYP2D6, CYP2C19, CYP2C9, CYP3A4, and CYP3A5 involved in the biotransformation of R- and S-fluoxetine to their N-desmethyl metabolites [65,66]. It was also demonstrated that CYP2C9 catalyzes R-fluoxetine demethylation and the formation of S-norfluoxetine is highly dependent on CYP2D6 [65,67]. Their catalytic activities depend on genetic polymorphisms. Moreover, fluoxetine has demonstrated inhibitory potency toward CYP2C19, CYP2C9, and CYP3A4, consequently having the potential to alter metabolism and pharmacokinetics of coadministered drugs metabolised through the same pathway or its own metabolism.
Despite the data provided by some studies, serotonin toxicity phenotype versus genotype association still needs more evidence-based research data. In the described case, the phenotype of ST was likely a consequence of variation in Cytochrome P450 enzyme activity caused by genetic variations in CYP genes. Cytochrome P450 isozymes are a major determinant of the pharmacokinetic behaviour of psychotropic drugs. This case demonstrated that responses to fluoxetine, even at therapeutic doses, can vary significantly between individuals. Similar dosages can have divergent results due to polymorphism in the genes that code for the isozymes responsible for the metabolism of drugs. Drug treatment response can be influenced by several factors, such asage, organ function, environmental factors and the nature and severity of disease, however, genetic polymorphisms account for most variations in treatment responses. The early diagnosis of genetically predisposed deficiency, based on molecular genetic methods, will lead to early recognition of serotonin toxicity and early intervention.

©2017 Piatkov, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.