Journal of
eISSN: 2373-4469


Research Article Volume 3 Issue 3
Department of Biotechnology, Haldia Institute of Technology, India
Correspondence: Pranab Roy, Department of Biotechnology,Haldia Institute of Technology, Haldia, 721657, India, Tel 91-9933037099
Received: November 08, 2016 | Published: December 28, 2016
Citation: Roy P, Guha D, Banerjee R, et al. A comparative study of three rhizospheric bacteria belonging to different genera, co-infecting a leguminous plant. J Investig Genomics. 2016;3(3):63-73. DOI: 10.15406/jig.2016.03.00053
Nodules obtained from the roots of fenugreek (Trigonella foenum graecum), a leguminous plant yielded three distinct classes of Nitrogen fixing bacteria. Molecular phylogeny based on 16S rRNA gene sequences showed them to be Enterobacter cloacae (R1), Pantoea dispersa (R10) and Enterobacter ludwigii (R12). The biochemical and morphological characterization of the three bacteria showed many similarities. All were gram negative, rod shaped, giving identical biochemical reactions like Catalase test, MR tests, oxidase test etc. However there were differences in their sensitivity to the abiotic stresses like salinity, tolerance to heavy metal ions and temperature. Sensitivity to antibiotics also showed similar responses. Co-infecting the same plant and occurrence in the same nodules by different genera of bacteria, capable of Nitrogen fixation is a novel phenomenon. The commonly held belief that all Nitrogen fixing mucoid bacteria occurring in nodules of legumes are Rhizobial species is not tenable. Plant growth promoting effects of these bacteria were observed by IAA, siderophore production and inhibition of ethylene biosynthesis by plants by producing the enzyme, ACC Deaminase. The host specificity for one of the bacterium, R1 was shown as only Fenugreek showed nodulation but not mung bean or Bengal Gram seedlings. Nitrogen fixation by the isolated bacteria showed all of these could reduce acetylene to ethylene by gas chromatography.
Keywords: Nitrogen fixation; Nodules; Enterobacter species; Co-infection; Gas chromatography; Rhizobial
Plant-microbe symbiosis has been recognized as the major source of Nitrogen supply to leguminous plants. The root nodules of most of the legumes are associated with Rhizobium sp. and Bradyrhizobium species, which can fix atmospheric Nitrogen gas into ammonia and further to nitrite and nitrate [1,2]. Recent studies have shown that there are many other genus and species of bacteria, associated with root nodules which can effectively fix Nitrogen, provide the host plant with growth hormones like IAA, reduce ethylene production by synthesizing the enzyme ACC Deaminase and produce siderophores to chelate insoluble metal ions like Fe (III) and make them available to the host plant [3]. In order to find novel Nitrogen fixing micro-organisms from leguminous root nodules, which are more tolerant to higher temperature and salinity, we chose Fenugreek as the plant hosting the nodulating microbes. In our studies, we isolated and characterized three novel bacteria from the root nodules of Fenugreek (Trigonella foenum). Though belonging to different genera like Enterobacter and Pantoea, these bacteria shared many characters like gram staining, antibiotic sensitivity, IAA production, siderophore production, motility etc; however, there were differences in tolerance to salinity, heavy metal ions and temperature. Pantoea species have been isolated from a wide variety of hosts, both plants and animals; this was classified as Enterobacteriaceae family [4]. This study revealed that a single leguminous plant root can be nodulated by different types of bacteria, fixing Nitrogen and helping the plant by providing resources.
Viable seeds of Fenugreek, Mung bean and Bengal grams were obtained from the local suppliers. All chemicals used in this study are of AR grade, purchased from Himedia and SRL. Acetylene reduction assay for biological nitrogen fixation was carried out at University of Calcutta, West Bengal. All the assays were performed thrice and the standard errors of mean are shown in the graphs.
Isolation and characterization
Nodules of Fenugreek (Trigonella foenum-graecum) were taken from field grown plants , surface sterilized with 10% sodium hypochlorite followed by 70% ethanol wash and several washes with sterile water. Nodules were then crushed and suspended in phosphate buffered saline. Several dilutions were prepared and plated on CRYEMA (Congo Red, yeast extract, mannitol agar) and individual colonies were isolated. Three such colonies, named R1, R10, R12 were further studied.
Biochemical tests, morphology and motility were determined for each isolate. Gram staining was done for the three isolates for their characterization using the standard protocol.
Molecular identification of the three isolates
Genomic DNA was isolated and tested on 0.8% agarose gel electrophoresis. The 16S rRNA genes were amplified by using the universal forward primer (27F) and the reverse primer (1492R) under standard PCR conditions. The sequencing of the 1.4kb amplicons were carried out by automated DNA sequencer at Chromous Biotech Pvt. Ltd., Bangalore, India. The sequences were deposited at NCBI with the accession numbers, KX68756 (R1), KX687557 (R10), KX687554 (R12). Molecular phylogenetic analysis was done with MEGA 7.1 software.
Bacterial growth curve
24 hour grown fresh cultures of the three isolates were taken and inoculated of fresh LB broth and the bacterial growth pattern was studied at 30˚C spectrophotometrically from 0 to 14 hours by taking absorbance at 650nm at every one hour. Graphs were plotted with bacterial growth against time intervals in hours.
Root infection test of the R1 isolate
Plant infection and root nodule formation confirm the host specificity of a particular bacterium. This study also provides the basis to screen the efficiency of a bacterial strain for nodulation and nitrogen fixation.
Viable seeds of Fenugreek (Trigonella foenum-graecum) (M), Mung bean (MB) and Bengal gram (BG) were obtained from local suppliers. Seeds were surface sterilized with 10% sodium hypochlorite followed by 70% ethanol wash and several washes with sterile water. Seeds were soaked in water overnight for germination. Seeds were placed in Murashig Skoog (MS) medium in choir bed in different sets, Set-I contained MS medium without Nitrogen (N) source and with the R1 inoculum, Set-II contained MS medium with N source but was without the inoculum and Set-III had both MS medium with N source and the bacterial inoculums. Seedlings were kept in Plant chamber maintaining 16 hrs light and 8hrsdarkness for about 28 days. The roots of the seedlings were observed and studied after 15th day and 28th day under microscope.
Protein estimation from the root extracts of the seedings
Protein was estimated from the root extracts by Bradford [5]. Root extracts were crushed and the extracts were suspended in phosphate buffer saline. A protein standard curve was prepared with BSA(1mg/ml). Extracts were then incubated for 5-15 mins with the Bradford reagent until the blue colour develops and then the absorbance was recorded at 595nm.
Carbohydrate esitmation from the root extracts of the seedlings :
Carbohydrate was estimated from the root extracts by the phenol sulphuric acid method [6].
Salinity tolerance
Salinity tolerance of the three isolates were studied in LB (Luria-Bertani) medium with different concentrations of NaCl (0 to 1M).
48 hours cultures were taken and inoculated in LB medium with varying salt concentrations and incubated at 30˚C for 24 hours in shaker and absorbance was taken at 600 nm [7].
Temperature tolerance
Temperature tolerance of the three isolates were studied by plating on LB agar and incubating for 24 and 48 hours at different temperatures 30˚C, 37˚C, 42˚C. Colonies were observed to find out their tolerance to different temperatures.
Nitrogen fixation
All the three isolates were capable of growing in Nitrogen free media like Jensen’s agar and YEM agar, indicating their ability to fix atmospheric Nitrogen. The three isolates were tested by the Acetylene reduction assay where acetylene is reduced to ethylene by the enzyme Nitrogenase, responsible for nitrogen fixation. Ethylene peaks measured by Gas chromatography were quantified for the enzyme activities [8].
Antibiotic sensitivity of the three isolates
Antibiotic sensitivity was observed by spreading the three purified cultures on LB plates on which different antibiotic containing filter discs were placed and incubated overnight at 30˚C. The antibiotics used in this study are: Ampicillin (25µg/disc), Gentamycin (10µg/disc), Trimethoprim (30µg/disc), Tetracycline(30µg/disc), Streptomycin (10µg/disc), Vancomycin (30µg/disc), Doxycycline (30µg/disc), Cloxacillin (10µg/disc) The zone of inhibition around the filter discs (from Himedia) were measured in diameter. The sensitivity and resistivity of the three isolates with each antibiotic were observed.
Heavy metals tolerance
Heavy metal ions tolerance was studied by making stock solutions of 10mM each and then broth cultures of the three isolates were inoculated on fresh LB medium supplemented with metal ions (Pb,Cr,Hg,Co) of increasing concentrations (83,166,250,330,410,500µM) followed by incubation at 30˚C overnight. The absorbance was recorded at 600 nm [9].
Indole acetic acid production
Indole-3-acetic acid (IAA) is a phytohormone, which is essential for the growth and development of plants. It is known to increases in cell elongation and cell division and differentiation responses in plants [10,11]. The capacity to synthesize IAA is widespread among soil- and plant associated bacteria. It has been estimated that most of the bacteria isolated from the rhizosphere can produce the plant growth regulator IAA . Tryptophan is the precursor of IAA [12].
The colorimetric technique proposed by Gordon et al. [13], the three isolates were grown LB liquid medium with tryptophan (1mg/mL) at 30˚C with shaking for three days. The supernatant of the culture was recovered after centrifuged. Thereafter, supernatant was taken in a test tube and 2 mL IAA reagent (1 mL of 0.5 M FeCl3 was mixed in 50 mL of 35% HClO4) was added. After incubation for 20 min at room temperature, the optical density of the samples was recorded at 550 nm with respect to blank medium as the control.
ACC Deaminase assay
The ACC Deaminase activities of the isolates were qualitatively measured in agar plates following the method of Glick et al. [14]. The isolates were inoculated on solid medium plates containing carbon and nitrogen free DF salt minimal medium [15] supplemented with ACC where ACC act as sole carbon and nitrogen source. The plates were incubated at 30˚C for 3 days and observed for colony formation. The colonies formed in DF medium supplemented with ACC were considered positive for ACC Deaminase activity. DF salt minimal medium without ACC supplement was used as control. The quantitative estimation of ACC Deaminase activity was assayed according to a modified method of Honma et al. [3], which measures the amount of α-ketobutyrate produced when the ACC Deaminase cleaves ACC. The amount of α-ketobutyrate was quantified spectrophotometrically at 550 nm.
Detection of siderophore production
Assay for siderophore production by the three isolates was carried out by spot inoculating purified cultures on Chrome azurol S agar plates and incubated at 30˚C for 3-4 days [16].
Molecular identification and characterization
Bacterial isolates R1, R10 and R12 colonies were all mucoid with exopolysaccharide. 16S rRNA sequencing identified the three isolates as, isolate R1 to be Enterobacter cloacae [17], isolate R10 to be Pantoea dispersa and isolate R12 to be Enterobacter ludwigii (Table 1).
Sample Code |
Source |
Accession Numbers |
Identified Organisms |
R1 |
Root nodules |
KX687556 |
Enterobacter cloacae |
R10 |
Root nodules |
KX687557 |
Pantoea dispersa |
R12 |
Root nodules |
KX687554 |
Enterobacter ludwigii |
A1 |
Rhizospheric soil |
KX687553 |
Pseudomonas sp |
P1 |
Rhizospheric soil |
KX687555 |
Pseudomonas sp |
Table 1: Molecular identification and characterization.
The phylogenetic trees for the three isolates generated by the software MEGA 7.1 were obtained from the sequence data of the 16S rDNAs as follows: (Figure 1A-1C).
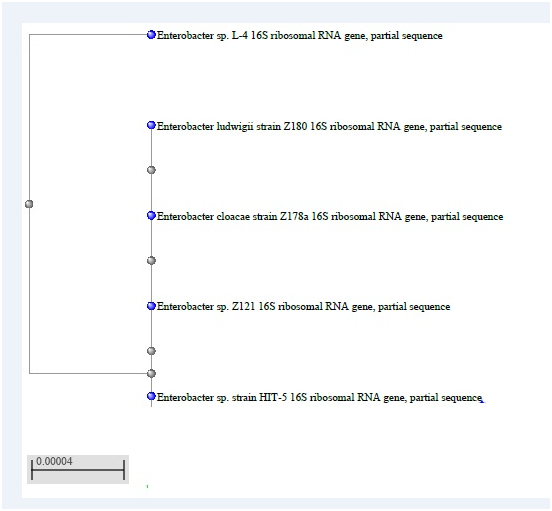
1A: HIT 5 = R12 Isolate.

1B: R1 Isolate.

1C: R 10 Isolate
Gram staining
All the three isolates were found gram negative short rods under 100X magnification (Figure 2).

2A:R1 Isolate.

2B: R12 Isolate.

2C: R10 Isolate.
Biochemical tests
The routine biochemical tests for each of the three isolates were carried out (Table 2). Many of the tests gave identical results except VP test where R10 isolate showed positive result where the other two were negative and in Indole test R12 isolate showed positive result where the other two were negative. TSI agar test showed R10 isolate to be different from R1 and R12 isolates. Since both R1 and R12 were Enterobacter species, it showed similar reactions for metabolizing different sugars, but R10, a Pantoea specie was different.
Test |
R1 |
R10 |
R12 |
1.Catalase |
- |
- |
- |
2.Citrate |
++ |
++ |
++ |
3.Oxidase |
++ |
++ |
++ |
4.Indole |
- |
- |
++ |
5.MR |
- |
- |
- |
6.VP |
++ |
- |
- |
7.Gelatinase |
- |
- |
- |
8. Growth on Glucose peptone agar |
++ |
++ |
++ |
9.Motility |
+++ |
+++ |
+++ |
10.TSI Agar test |
Acid formation with no gas formation and all the three sugars were fermented |
Acid formation with gas formation and only glucose fermented |
Acid formation with no gas formation and all the three sugars were fermented |
Table 2: The routine biochemical tests for each of the three isolates were carried out.
Root infection test of the R1 isolate
Figure 3 shows the root infection of Fenugreek seedling. Only Fenugreek showed nodulation after 28 days with R1 inoculation showing host specificity. Only Fenugreek roots were nodulated by R1 but not mung bean or Bengal gram. Figure 4 shows the initiation of the nodule formation in Fenugreek seedling after 15 days of infection with R1 isolate.
Protein and carbohydrate estimation from the root extracts
Figure 5A & 5B for carbohydrate and protein estimation with seedlings of Fenugreek, Mung bean and Bengal gram. Set III which had N source and inoculated with R1 had the highest levels of protein and carbohydrate for all the three seedlings.

5A

5B
Bacterial growth curve
Figure 6a-6c show the growth kinetics of the three isolates, where the absorbances of the cultures at different time points after inoculation are plotted against time in hours. The growth rates of all the three were very similar in LB medium at 30˚C.

6A: R1
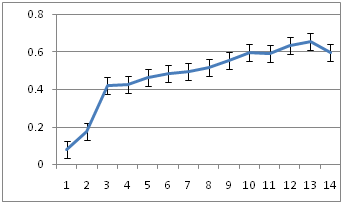
6B: R10
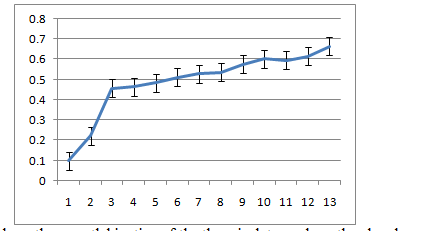
5C: R12
Salinity tolerance
Figure 7a-7c shows the bacterial growth in different salt concentration and their tolerance towards salt stress. Isolate R12 (Figure 7c) shows 50% inhibition at 500mM NaCl salt concentration and proves to be highly salt tolerant whereas isolate R10 (7b) and isolate R1(7a) shows 50% inhibition at 50 mM and 350 mM NaCl salt concentrations respectively. Thus the salinity tolerances of the three nodule associated bacteria were quite different.
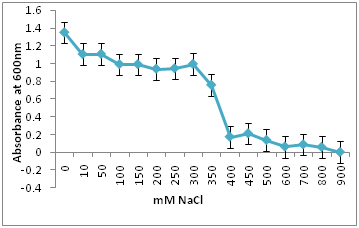
7a. R1 Isolate: Isolate R1 shows 50% inhibition at 350 mM NaCl salt concentrations.

7b. R10 isolate: Isolate R10 shows 50% inhibition at 50 mM NaCl salt concentrations.

7c.R12 isolate: Isolate R12 shows 50% inhibition at 500mM NaCl salt concentration and proves to be highly salt tolerant.
Temperature tolerance
Table 3 shows isolate R1 and R12 had optimum growth at 37˚C whereas isolate R10 grew maximally at 30˚C. Temperature tolerance of the three isolates were compared in LB medium. Both the Enterobacter species, R1 and R12 showed maximum growth at 37˚C whereas R10 (Pantoea) which is reported to be cold tolerant had better growth at 30˚C [18].
Bacterial Species |
Temperature |
||
30˚C |
37˚C |
42˚C |
|
1. Enterobacter cloacae (R1) |
+++ + |
+++ +++ |
++ |
2. Pantoea dispersa (R10) |
++++++ |
++ |
+ |
3. Enterobacter ludwigii (R12) |
+++ |
+++ +++ |
+ |
Table 3: Shows isolate R1 and R12 had optimum growth at 37°C whereas isolate R10 grew maximally at 30°C.
Nitrogen fixation
Acetylene Reduction Assay was carried out with the three isolated colonies on agar slants using Gas Chromatography. R12 showed the highest activity followed by R1 and R 10 respectively as calculated by the areas under the peaks of ethylene in the chromatograms (data not shown).
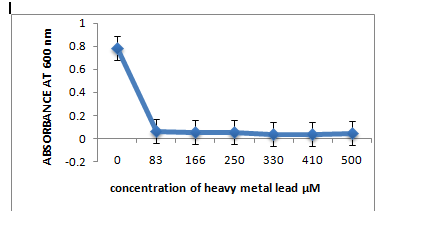
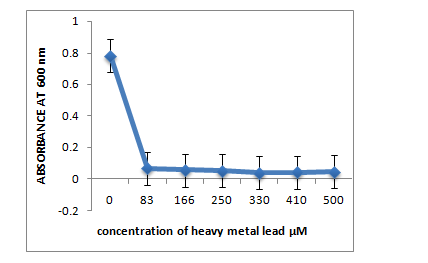
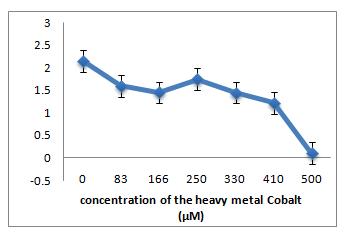
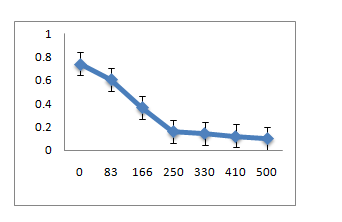
Metal ion toxicity
The three strains R1, R10 and R12, grown in LB medium with different concentrations of metal ions Pb(II), Co(II), Cr(VI), Hg(II) showed that R1 strain was tolerant to Pb and Co whereas it was sensitive to Cr and Hg ions at 83µM. R10 and R12 strains were sensitive to Pb but more resistant to Co, Cr and Hg at 83µM concentration.

A: R1 Isolate: (Enterobacter cloacae)
B: R10 Isolate: (Pantoea dispersa).
C: R12 Isolate: (Enterobacter ludwigii)
A. R1 Isolate: (Enterobacter cloacae)
B. R10 Isolate: (Pantoea dispersa)
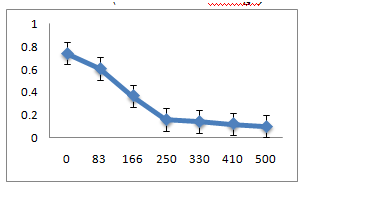
C. R12 Isolate: (Enterobacter ludwigii)
Antibiotic sensitivity
Isolates R1, R10 and R12 showed identical results on antibiotic discs when placed on solid media. R1, R10 and R12 were found to be sensitive to Ampicillin, Vancomycin and Cloxacllin antibiotics whereas showed resistance to all the other antibiotics (Table 4).
Antibiotics |
R1 |
R10 |
R12 |
Ampicillin |
Sensitive |
Sensitive |
Sensitive |
Gentamycin |
Resistant |
Resistant |
Resistant |
Trimethoprim |
Resistant |
Resistant |
Resistant |
Tetracycline |
Resistant |
Resistant |
Resistant |
Streptomycin |
Resistant |
Resistant |
Resistant |
Vancomycin |
Sensitive |
Sensitive |
Sensitive |
Cloxacillin |
Sensitive |
Sensitive |
Sensitive |
Table 4: Antibiotic sensitivity.
Indole acetic acid production
The three isolated bacteria R1, R10 and R12 were able to produce indole acetic acid when recorded spectrophotometrically whereas a negative control E. coli did not produce IAA (Table 5).
R1 |
R10 |
R12 |
|
++ |
+++++++ |
++++ |
|
Table 5: Indole acetic acid production.
ACC Deaminase activity
The three isolates were found to have ACC Deaminase activity when plated on DF minimal media supplemented with ACC as the sole carbon and nitrogen source. Colonies appeared on the plates; also colorimetrically it had been shown to grow in DF medium whereas a negative control showed no growth in this minimal medium. ACC Deaminase is the enzyme which reduces the biosynthesis of ethylene, a plant hormone inhibiting plant growth. Thus these bacteria act as plant growth promoters (Table 6).
R1 |
R10 |
R12 |
+++ |
+++++ |
+++++++ |
Table 6: ACC Deaminase activity.
The three isolates showed growth on Chrome Azurol S agar plates forming orange halo zone around the colonies against the blue background. Half of the plates were streaked with individual bacterium and the other half was spotted with the same. This indicates that the three isolates produced siderophores which can chelate Fe(III) from insoluble to soluble form, making it available to plants (Figure 8).
Amongst the Bio fertilizers used commercially Nitrogen fixing microbes and Phosphate solubilizing bacteria are the most common. The performance of these bacteria under unfavorable climatic conditions are poor. High temperature, salinity and occurrence of heavy metal ions inhibit the activities of these micro-organisms. In order to study the microbes associated with a leguminous plant root, we isolated three novel bacteria from Fenugreek root nodules. The biochemical and morphological characterization followed by molecular identification of the three isolates were carried out. These were found to be R1 (Enterobacter cloacae), R10 (Pantoea dispersa) and R12 (Enterobacter ludwigii) respectively. Antibiotics sensitivity and plant growth promoting activities were found to be similar in all three.

©2016 Roy, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.