Journal of
eISSN: 2373-6453


Research Article Volume 7 Issue 2
Institute of Progressive Medicine, South Pasadena, USA
Correspondence: W. John Martin, Institute of Progressive Medicine, South Pasadena, 1634 Spruce Street South Pasadena CA 91030, USA
Received: December 05, 2019 | Published: December 11, 2019
Citation: Martin WJ. Renegade cellular and bacterial genetic sequences in monkey-derived stealth adapted viruses. J Hum Virol Retrovirology. 2019;7(2):26-40. DOI: 10.15406/jhvrv.2019.07.00211
Stealth adapted viruses differ from the viruses from which they are derived in not being effectively recognized by the cellular immune system. This is because of the deletion or mutation of the genes coding for the relatively few virus components, which are generally targeted by cytotoxic T lymphocytes. Stealth adapted viruses do not, therefore, normally evoke inflammation, the hallmark of most infectious illnesses. A stealth adapted virus was repeatedly cultured from the blood of a patient with the chronic fatigue syndrome (CFS). Polymerase chain reaction (PCR) performed on the culture identified the virus as being derived from an African green monkey simian cytomegalovirus (SCMV). The PCR also amplified a genetic sequence closely related to a normal cellular gene. Further analysis of the viral DNA indicated that it was fragmented and genetically unstable. Moreover, additional genetic sequences have been incorporated into the replicating virus genome. Several of the additional sequences are originally of cellular origin with subsequent genetic modifications. Other incorporated sequences are of bacteria origin. PCR performed on cultures from some other CFS patients, led only to the amplification of modified cellular sequences, including a sequence apparently derived from the rhesus monkey genome. It is proposed that as part of the stealth adaptation process, sequences of the original infecting virus can be largely displaced by cellular and/or bacteria sequences, which have essentially switched their affiliation to that of the stealth adapted virus. For this reason, they are referred to as renegade sequences. The term “renegade viruses.” is also proposed to describe those viruses in which the originating conventional virus sequences have yet to be detected. The findings are relevant to efforts to seek a virus cause of many common illnesses, including CFS, and to the possible misattribution of certain illnesses to bacterial infections.
Keywords: renegade viruses, stealth adapted viruses, chronic fatigue syndrome, polio vaccine, noncoding rna, ochrobactrum, mycoplasma, cytomegalovirus, viteria
ACE, alternative cellular energy; CFS, chronic fatigue syndrome; CPE, cytopathic effect; CFS, cerebrospinal fluid; NCBI, national center for biotechnology information; NT, nucleotide; PCR, polymerase chain reaction; RhCMV, rhesus monkey cytomegalovirus; SCMV, African green monkey simian cytomegalovirus
A stealth adapted virus was repeatedly cultured from a CFS patient.1 It induces a foamy vacuolated cytopathic effect (CPE) when cultured on human and animal cells, including human foreskin fibroblast cells (MHRF). Electron microscopy of the infected cells indicate the presence of both herpesvirus-like particles and intracellular inclusions. The polymerase chain reaction (PCR) was employed in an effort to identify the type of virus infecting the cells. Among the sets of primers used in the PCR were a forward primer “SK43” (5’-cggatacccagtctacgtgt-3’), which matches to sequence within the tax gene of human T lymphotropic virus-1 (HTLV-I) and a backward primer “SK44” (5’-gagctgacaacgcgtccatcg-3’), which matches to a sequence within the tax gene of HTLV-II.2 Using relatively low stringency PCR, this set of primers yielded several PCR products when performed on the infected cultures, with no identifiable products being formed when using uninfected cultures.1 The PCR products were cloned and sequenced. A radiolabeled PCR product was also used to identify virus DNA in extracts of infected cells and in the pelleted material obtained by ultracentrifugation of filtered culture supernatant. In agarose gel electrophoresis, the virus DNA in the pelleted material migrated with an estimated size of approximately 20 kilobases (kb).1
Cytomegalovirus-related DNA sequences
The PCR assay yielded prominent bands in agarose gel electrophoresis of approximately 0.5 and 1.5kb.1 The 1.5 kb band was subsequently shown to contain two PCR products, measuring 1,487 and 1,510 nucleotides, respectively. The PCR amplified sequences were flanked at both ends by the SK44 primer. Using the BLASTN and BLASTX programs 3, the amplified sequences were compared with the then available genetic sequences on GenBank; a repository of nucleic acid and protein sequences maintained by the National Center of Biotechnology Information (NCBI). The analysis showed that the longest sequences (included in NCBI Accession number U09212) was related to but still different from a sequence within human cytomegalovirus. As more GenBank sequences became available for comparison, the sequence of this PCR product was shown to have far greater sequence homology with African green monkey simian cytomegalovirus (SCMV).4 Similarly, the other large PCR product (NCBI Accession number U09213) was subsequently shown to match closely to a sequence within SCMV with 1348/1454 (93%) identical nucleotides, with 19 gaps.5 The next closest match of the sequence of the PCR product is to the cytomegalovirus of Mandrillus (Dril) monkey with 938/1389 (68%) identical nucleotides and 124 gaps. There was even less overlap with the best matching rhesus monkey cytomegalovirus (RhCMV,) which had 364/515 identical nucleotides. Using the standard BLASTN program, there was no significant matching of the sequence in U09213 with human cytomegalovirus.
Detection of cellular-derived DNA sequence
The smaller product that was amplified in the PCR on infected but not on uninfected cultures was sequenced (NCBI accession number AF107851.1). This product was flanked at both ends with primers in which there was a single nucleotide difference (cytosine rather than adenosine at the 10th position) than in the originally intended SK43 primer. The DNA sequence of the PCR31-637 amplified product, excluding the primers, was analyzed against the most recent “assembled” complete human genome, This showed a near identical homology with an intergenic region of the human X chromosome (NCBI accession number NC_000023.11), extending from nucleotide 30,177,214 to nucleotide 30,177,822. Within this region there were 606/609 identical nucleotides (99.51 percent identity) with 2 single nucleotide deletions and one nucleotide substitution (Figure 1).

Figure 1 Matching of the sequence of the PCR product with that of a region of the human X chromosome
When using the nr/nt (protein and nucleotide) database in the BLASTN program, there were only two matches to the human genome (NCBI Accession numbers AC117518.3 and AC005145.1). The next 3 matches, all with 97.5% identity, were to a sequence in the chimpanzee genome. It was considered possible that the X-chromosome-related sequence may have originated in an African green monkey. On further analysis using the primates’ “assembled genomes,” there was decreasing overall relatedness of the PCR product with the human genome to the genomes of chimpanzee, rhesus, and Cercopithecus (African green) monkeys. The data are included in Table 1 and establishes the X-chromosome related genetic sequence as being human in origin.
|
Species |
NCBI accession no. |
Identical nucleotides |
% Identity |
Gaps |
|
Human |
NC_000023.11 |
606/609
|
99.51 |
2 |
|
Chimpanzee |
NC_036902.1 |
598/609
|
98.19 |
2 |
|
Rhesus monkey |
NC_041774.1 |
555/609
|
91.13 |
5 |
|
African green monkey |
NC_023671.1 |
553/609 |
90.80 |
2 |
Table 1 The relative number and percentages of identical nucleotides of the cloned PCR product shown in Figure 1 with matching sequences of humans and several primates
Cloning of the virus genome
Filtered supernatant of infected cultures was ultracentrifugation as a means of pelleting extracellular virus particles. DNA was extracted from the pelleted material. In a second study, the extracted DNA was further purified by electrophoresis into agarose gel, with excision of the narrowly banded DNA. The DNA from the first study was cut using the EcoR1 restriction enzyme and cloned into pBluescript plasmids (3B series). The DNA in the second study was cut using the Sac1 restriction enzyme and was also cloned into pBluescript plasmids (C16 series). At least partial DNA sequence data were obtained from over 450 clones using the T3 and T7 promoter sites on the pBluescript plasmid.5–7 Several of the more interesting clones were completely sequenced. The sequence data on the individual clones were provided to GenBank under the listing of stealth virus-1. Using the BLASTN and BLASTX programs of the NCBI, the sequences fell into three categories. The majority of the clones contained sequences, which matched reasonably closely to regions of the SCMV genome. Several of the clones with sequences that match to the SCMV genome have been assembled into longer stretches of non-overlapping sequences. The NCBI accession numbers of these longer stretches are listed in Table 2 and collectively they comprise 97,780 nucleotides. The overall regions of the SCMV genome (length 226,205 nucleotides) which the BLASTN program matches to the stretches of the stealth adapted virus sequences are shown in Table 2, along with the number of identical nucleotides. With sequences, U27627.2 and U27770.2, there was an intervening region in which the stealth virus and SCMV sequences did not adequately match. There are disproportionate numbers of clones, which match to certain regions of the SCMV while there is no matching to other regions. There is also diversity in the sequences in some of the clones, which match to the same region of the SCMV genome. These differences include nucleotide substitutions, deletions, repetitions, and recombination. Nevertheless, the results are unequivocal in indicating that SCMV was the originating virus and that it has undergone substantial genetic changes.
|
Accession number |
Length |
Overall region in SCMV with matching NT |
Number of identical NT in longest matching segment |
|
U09213.1 |
1,437 |
27,815 to 29,266 |
1,346/1,454 |
|
U27469.2 |
28,199 |
36,361 to 64,456 |
26,523/28,245 |
|
U27883.2 |
8,407 |
66,002 to 74,405 |
8027/8414 |
|
U27471.2 |
6,328 |
95,026 to 101,313 |
5,112/6367 |
|
U27238.2 |
25,023 |
111,877 to 136,936 |
23,941/25,092 |
|
U27627.2 |
12,375 |
149,167 to 166012 |
6,222/6654 |
|
U27770.2 |
16,011 |
202,123 to 218,661 |
11/583/12,242 |
Table 2 BLASTN Matching of the Cloned Sequences of Stealth Virus-1 with the sequence of Cercopithecine herpesvirus 5 strain 2715, NCBI FJ483968.2
Detection of additional cellular sequences related to the human genome
A second category of cloned sequences were those that could be matched to various specific regions of the human genome.8,9 The matching cellular sequences are located in different human chromosomes. While the sequences of some of the clones are identical to the matching region of the human genome, many of the clones showed minor differences. The most common nucleotide changes are multiple inserts of single nucleotides into the different cloned sequences. Occasionally longer inserts are present. The overall frequency of inserted nucleotides in the clones, which were different from the normal human sequence, is approximately 0.5% (1 in 200). Another change, also occurring in some of the clones with a frequency of approximately 0.5%, is single nucleotide substitutions. Three examples of comparing cloned sequences with those of the matching cellular sequence are provided in Figure 2. It is of course realized that some errors can occur in sequencing, It was also thought possible that some of the clones might possibly match better to the genome of African green monkeys. The sequence analysis was, therefore, occasionally repeated using the assembled genome of African green monkey. In no case so far examined, however, is the matching to the monkey genome been closer than that to the human genome. Another observation is that the regions of partial homology between the cloned sequence and that of a human cellular gene are invariably occurring within the non-coding region of the human genes. A more comprehensive analysis of the relatedness of the cloned sequences to the human and primate genomes is ongoing and will be provided in a subsequent publication .
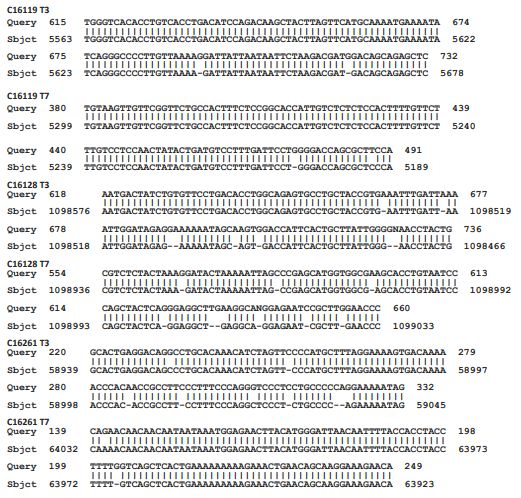
Figure 2 Matching of cloned sequences with cellular sequences.
What is shown are three examples in which a region from both the T3 and the T7 partial readouts of the sequences of three clones obtained from the purified DNA from the culture supernatants is directly compared with a matching Sbjct region of cellular sequences. The six selected cloned sequences are labeled as Query, whereas the Sbjct is the matching cellular sequences. The numbers refer to the numbered nucleotides in the clones and in the Sbjct. The Sbjt for clones C16119 , C16128, and C16261 are Homo sapiens chromosome 8 clone ABC9_45365100_H16, complete sequence AC275379.1; Homo sapiens kazrin, periplakin interacting protein (KAZN), RefSeqGene on chromosome 1 Sequence ID: NG_029844.2; and Homo sapiens isolate fa0190 immunoglobulin superfamily member 4B (IGSF4B) gene AY663433.1, respectively.
Detection of bacteria-derived sequences
The third category of sequences identified in the cloned DNA are clearly bacterial in origin 10. Even at the initial time of submission of the sequence data to GenBank, it was apparent that several of the clones were very closely related to mycoplasma bacteria.1. Examples of clones with near-identical mycoplasma-related sequences are 3B35, 3B512, 3B528, and 3B632. The sequences these clones closely matched to different sequences, which are essentially identical in plasmid 7 of Mycoplasma conjunctivae strain NCTC10147 and in plasmid 9 of mycoplasma fermentans strain M64.12, 13 The numbers of identical nucleotides in these four cloneswith the corresponding sequences in the mycoplasma plasmids 2136/2142, 2342/2345, 2020/2044, and 1380/1384 respectively. The sequences of several additional clones still preferentially match to mycoplasma but with lower levels of nucleotide identity.
An even larger number of the clones have sequences, which at the time of submission to GenBank, matched most closely to Brucella, an alphaproteobacterial.14 These sequences have subsequently been shown to have greater homology to one or other species of Ochrobactrum.15 Examples of these clones are NCBI accession numbers 3B23, 3B41, 3B43, 3B47, 3B534, 3B614, C1616, and C16134.The different cloned sequences tend to preferentially match to O. quorumnocens, with somewhat less matching to O. pituitosum and O. pseudogrignonense.16–18 Yet for some of the clones, there was only minimal or no matching to O. quorumnocens. Some of these data are summarized in Table 3. For many of the cloned sequences, the changes from the best matching Ochrobactrum species are rather minor with less than 1% nucleotide substitution and only occasional gaps. Still other clones have sequences, which, although seemingly of bacterial origin, cannot be currently assigned to a particular species of bacteria. Some of these clones appear to be broadly related to certain alphaproteobacteria, but only when using the BLASTX program of the NCBI. An example is clone 3B513 (NCBI accession number U27894.2), which comprises 8,106 nucleotides. Other cloned sequences of apparent bacteria origin show a weak association with Actinobacteria.
|
Clone |
O. quorumnocens |
O. pituitosum |
O. pseudogrignonense |
|
3B23
|
8911/8916(99%) |
7550/8955(84%) |
7531/8953(84%) |
|
3B41
|
93/261(74%) |
2184/2914(75%) |
2330/2886(81%) |
|
3B47
|
no matching |
no matching |
1449/2015(72%) |
|
3B313
|
7973/7985(99%) |
6639/7992(83%) |
6638/7992(83%) |
|
3B513
|
no matching |
no matching |
no matching |
|
3B614
|
5060/5062(99%) |
2561/3014(85%) |
2541/3002(85%) |
|
C1616
|
4598/4626(99%) |
1685/2013(84%) |
1689/2013(84%) |
|
C16134
|
4127/4143(99%) |
1203/1423(85%) |
1202/1423(84%) |
Table 3 The relative matching of the cloned sequences from the DNA of the stealth adapted virus cultures with three species of Ochrobactrum bacteria
Genetically unstable and fragmented virus genome
As noted above, the sequences in many of the clones correspond to diverse regions of the SCMV genome. There were many examples of significant differences between clones even when they matched to the same region of the SCMV genome. These differences were attributed to genetic instability of the virus genome, including mutations, deletions, insertions, and recombination. Furthermore, the aggregate length of non-overlapping virus sequences far exceeded the estimated 20kb genome size based on the hybridization pattern seen in agarose gel electrophoresis. This result is consistent with a fragmented genome.19 It is also possible that some of the DNA fragments are held together via RNA, which can be subjected to breakage from environmental sources of RNases.
PCR assays on other stealth adapted virus cultures
The SK43/SK44 set of PCR primers was used in PCR assays on positive stealth virus cultures obtained from several other patients. A strongly positive culture was obtained from the cerebrospinal fluid (CSF) of a comatose patient with a 4-year history of a bipolar psychosis.20 The culture gave similarly sized PCR products as did the previous culture. Partial sequencing on one of the products from this culture confirmed a close homology to that of the earlier sequenced PCR product from the prototype SCMV-derived stealth adapted virus ((NCBI Accession number U09213). There were still significant minor differences between the sequences of the two viruses and with the corresponding region of SCMV. Several sets of PCR primers were also developed based on sequences in the cloned region of the prototype SCMV-derived stealth adapted virus. These primers gave positive results when tested directly on the CSF obtained from the comatose patient. A positive culture from the CSF of another CFS patient, gave a negative PCR in direct testing using these primers sets. Yet the culture did show a positive PCR for SCMV-related sequences when the RNA from the culture was first transcribed into DNA using reverse transcriptase PCR.
SK43/SK44 primers are sometimes able to generate discrete PCR products when tested on positive cultures from CFS patients. Three cultures yielded products of approximately 0.5 kb, together with some smaller products. The larger product from one of the cultures was cloned into pBluescript and fully sequenced. Several PCR products from the other two cultures were similarly cloned and partially sequenced from the T3 and T7 promotor sites. The cultures were designated A, B, and C. The PCR product from culture A (NCBI AF107850.1) was flanked on one end by the SK43 primer and on the other end by the SK44 primer. The 507-nucleotide sequence between the primers was analyzed using BLASTN. The top listed matching result was to Homo sapiens Rho guanine nucleotide exchange factor 10 (ARHGEF10), RefSeqGene (LRG_234) on chromosome 8 (NCBI accession number NG_008480.1). The matching region extended from nucleotide 88302 to nucleotide 88794 of the cellular gene. This is a non-coding region of the gene. Similar to the matching with two additional GenBank entries that cover the same region of chromosome 8, there are 447/505 identical nucleotides (89%) with 16/505 gaps. A major gap occurs because of a nucleotide stretch reading “acccccccccact” in the PCR product between nucleotides 276 to 288. The matching is shown in Figure 3.

Figure 3 Matching of cloned sequences from culture A with a cellular sequence.
What is shown is the BLASTN readout of the nucleotide matching of the “Query” sequence of a cloned PCR product from culture A with a sequence within Homo sapiens Rho guanine nucleotide exchange factor 10 (ARHGEF10) gene on chromosome 8 (NCBI accession number NG_008480.1). Note the 13-nucleotide insert into the cloned DNA sequence from nucleotide 276 to 288.
Cloning of the PCR products from the other two cultures, B and C, identified two clones from culture B (C1113 and C1132). and one clone from culture C (C1332), which all have sequences closely related to the sequence of the PCR product from culture A. These comparisons are shown in Figure 4. Although, these clones were only partially sequenced, they are clearly related to one another and also to the cellular sequence within the ARHGEF10 gene. Yet, even when comparing the limited sequences, the clones slightly different from each other (Figure 4). This even applies to the comparison of the sequences of the two related clones obtained from culture B. Of particular interest these two clones also include the insert that differentiated the clone from culture A from the human cellular gene. The insert was slightly changed in clone C1113 with an adenosine replacing one of the cytosine nucleotides (Figure 4). The sequencing of the matching clone from culture C did not extend to the region of the insert to determine whether or not it was present. Yet in other regions, this clone matched more closely to the clone from culture A than to the cellular sequence.

Figure 4 Alignments of the sequence of the cloned PCR product culture A with the sequences in cloned PCR products from cultures B and C.
What is shown are Query sequences from the T3 and T7 sequence readouts from two clones from culture B (C1113 and C1132) and one clone from culture C (C1322) aligned with the sequence of the PCR product cloned from culture A (Sbjct). The alignments did not include the SK43 and SK44 primers.
Four additional clones obtained from culture B and 6 additional plasmids obtained from culture C were partially sequenced using the T3 and T7 promotors on the pBluescript plasmid. Even though the cultures were obtained from different patients, there are two sets of matching clones. The T3 and the T7 sequences beyond the primer sites of Clone C1123 from culture B are similar to the sequences read from the T7 and the T3 promotors in clone C1313 from culture C (Figure 5) Similarly, the T3 and T7 readouts from clone C1142 from culture B are similar to the T3 and T7 readouts from clone C1335 from culture C (Figure 5). The remaining two clones from culture B and four clones from culture C were different from one another. By BLASTN analyses, the best matchings for three of the four clones from culture B and both of the clones from culture C, are each to different regions of the human genome. None of the matchings is greater than 50% nucleotide identity (data not shown). The remaining clone from culture B 5 of the clones They did, however, show partial matching to different regions of the human genome.


Figure 5 Alignment of matching clones from culture B and C.
What is shown are the alignments between the available T3 and T7 sequences of two sets of matching clones from culture B and culture C. These matching clones were in addition to the clones which matched to the cloned PCR product from culture A. The alignments show close similarity but not identity.
Rhesus Monkey-Related Cellular Sequence
Clone C1163 from culture B, NCBI accession number AF067480, comprises 90 nucleotides between the SK44 and SK 43 primers. By BLASTN analysis, these nucleotides exactly matched (100%) to a cellular sequence in a rhesus monkey (Figure 1). The identical matching was with MACACA MULATTA BAC clone CH250-195L5 (NCBI accession number AC242917.4). There was also exact matching with the two assembled rhesus genomes available on GenBank. There were 3 of 90 nucleotide differences between this clone and a region on chromosome 1 of the grivet African green monkey. The best matching to the human genome showed 10 of 90 nucleotide differences. (Figure 6), which was similar to the matching with the chimpanzee genome. The matching sequence in the rhesus monkey genome is intergenic since a BLASTX analysis of the matching and adjoining regions of the rhesus genome shows no identifiable coded proteins.
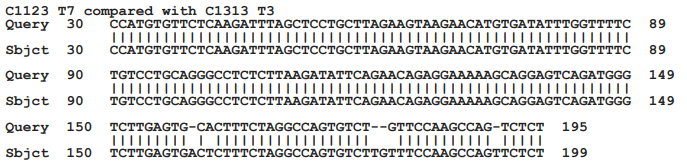

Figure 6 Matching of the sequence of the cloned PCR product with primate and human genomes.
What is shown is the sequence of clone C1162 from culture B as the Query, with the best matching cellular sequences (Sbjct) in the rhesus monkey, African green monkey and human genomes. The chimpanzee genome gave the same matching result as did the human genome.
These results provide additional information regarding stealth adapted viruses. In addition to a mechanism for evading cellular immunity, the process of stealth adaptation can apparently lead to the substitution of some and potentially all of the originating virus genome with other genetic sequences. The substituted sequences can be cellular and/or bacterial in origin. These substituted sequences, as well as any remaining initial virus sequences, can undergo further changes due to the fragmentation and genetic instability of the reformed viruses.
The incorporation of both cellular and bacterial sequences has occurred with an SCMV-derived stealth adapted virus.6–10 This virus is formally referred to as stealth virus-1. It was repeatedly cultured from a CFS patient. Low stringency PCR performed on virus infected cells using a fortuitous set of SK43/SK44 PCR primers leads to the amplification of two long SCMV-related nucleotide sequences and a shorter sequence with near identity to an intergenic region of the human X chromosome. No discernable PCR products are generated when the PCR assay is performed using the same set of primers on uninfected cells. The X chromosome related sequence was amplified through loose cross-reactions with the SK43 primer, which inadvertently had a cytosine rather than an adenosine as the tenth nucleotide. As was shown in Figure 1, the amplified sequence differs from the matching region in the human X chromosome in having two deleted nucleotides and one nucleotide substitution. The effective amplification of the sequence in the infected cells could possibly also be due to some minor nucleotide changes which would allow for more effective binding of the SK43 primer. This possibility can be addressed by further sequencing of the primer binding sites of the virus associated DNA. It is considered much more likely, however, that the amplification seen with the virus infected cells reflects a greatly increased copy number of the X chromosome-related sequence. This is consistent with the X-chromosome-related sequence having been incorporated into the virus replication process.
Several additional cellular derived sequences were identified upon cloning of the DNA isolated from the supernatant of the stealth adapted virus infected culture.6 In one of the two cloning studies, the DNA had been further purified by agarose gel electrophoresis prior to its restriction enzyme cutting and cloning into pBluescript plasmids. This was intended to reduce the possible inadvertent cloning of normal cellular DNA. Virus incorporated cellular DNA can also be distinguished from normal cellular DNA if it has undergone any mutations. Indeed, minor nucleotide differences from normal cellular DNA were seen in many of the sequenced clones with cellular-related DNA. As will be detailed in future publications, the DNA in these and in other cellular sequence-related clones will typically match to intragenic, and occasionally to intergenic, non-coding regions within a human genome.
The more common minor differences between the amplified sequences and their matching cellular sequence are i) added or deleted single nucleotides and ii) single nucleotide substitutions, each occurring with an overall frequency of approximately 0.5%. The sequences in several additional clones show far greater deviation from known cellular or viral sequences, sometimes with no convincing matching being obtainable to the human genome using the standard BLASTN program. This lack of precise matching is in spite of the human and many primate genomes having been fully sequenced. It is probably a reflection of the genetic instability of the sequences, including major insertions, deletions, substitutions, and recombination. To date, there has been no decisive example of contiguous SCMV and cellular sequences within the same clone. This type of recombination is, however, fully anticipated to be seen in further sequencing studies.
The incorporation of cellular sequences into stealth adapted viruses has also occurred in the stealth adapted viruses cultured in human cells from three other CFS patients. These cultures are designated A, B, and C. They were selected for further study because in low stringency PCR assays using the SK43/SK44 primers, they each generated a similar sized product as seen in agarose gel electrophoresis. A cloned PCR product from culture A has a sequence, which matches to a non-coding region within the Rho guanine nucleotide exchange factor 10 gene on human chromosome 8. Two of the cloned PCR products from culture B and one of the cloned PCR products from culture C also have sequences that match to the same human gene. As expected, the four PCR clones also match to one another. Yet the individual clones differ somewhat from each other, with one important exception. A cytosine rich 13 nucleotide sequence in the cloned PCR product from culture A is not present in the matching cellular gene. Yet the insert, one with a subsequent nucleotide substitution, is present in the two matching clones from culture B. Moreover, it cannot be excluded that the insert may have been present in the related clone from culture C The issue of possible cross contamination by a single clone is excluded since there are sequence differences elsewhere in the matching clones.
Rather it is indicative that a structurally altered human sequence is able to be transmitted between individuals. In other words, the sequence may not have originated in the patient’s own genome but has entered into the patient via a stealth adapted virus infection. The concept of transmissible cellular-derived genetic sequences is strengthened by the finding of two additional sets of similar clones in the PCR amplified materials from cultures B and C. Even more compelling for the infectious transfer of cellular-derived genetic material is the finding of rhesus monkey cellular-related sequence in one of the PCR products from culture B.
SCMV-derived stealth adapted viruses have presumably entered into the human population as a consequence of using SCMV infected African green monkeys to produce live polio virus vaccines.21,22 Earlier polio virus vaccines were produced in cytomegalovirus infected rhesus monkeys. DNA from Rhesus monkey cytomegalovirus (RhCMV) is present in some earlier polio virus vaccines, including the CHAT vaccine, which was experimentally tested in African chimpanzees.23 This is consistent with the report by Dr. Albert Sabin that the CHAT vaccine had a contaminating cytopathic virus.24 Indeed, it is likely that stealth adapted RhCMV was involved in the transformation of simian immunodeficiency virus (SIV) to human immunodeficiency virus (HIV).25
A reasonable model for the incorporation of cellular sequences into the virus genome is the hybridization of a fragment from the virus genome with a molecule of single stranded cellular RNA. This could lead to RNA or DNA synthesis of the complimentary sequence. If a different virus fragment then bound to the newly synthesized nucleic acid, it could essentially mimic the PCR within the cell. What is required, therefore, is a degree of genetic complementary between a fragment of the virus genome with a sequence in cellular RNA, together with an effective polymerase enzyme. Various cellular polymerases, including endogenous reverse transcriptase enzymes 26, can potentially be involved in the replication process. The polymerases of other viruses could similarly be involved. This is relevant to the production of polio vaccine virus in SCMV infected African green monkey kidney cells since the polio virus would be a source of RNA polymerase.27 Although, the cellular gene incorporation process for stealth virus-1 may have begun in African green monkeys, there may have been subsequent switching or exchanging of the incorporated primate cellular sequences with the related human cellular sequences. This type of exchange would evolve over time once humans became infected. The is consistent with the relatively close relationship of the PCR amplified sequence from the human X chromosome with the counterpart sequences in the genomes of primates. Stealth virus-1 induces a severe acute illness in cats.28 To test the possibility of genetic exchange, therefore, virus passaged in cats could be periodically examined for an exchange of the human sequences with feline sequences. Similar studies can be conducted in the many species of animal cells in which the stealth adapted virus can be cultured.1
The process by which cellular sequences have become part of the virus replication process can seemingly also apply to the incorporation of bacteria sequences. There is no evidence that the stealth adapted cultures were infected with any bacteria. Specifically, on several occasions, subcultures were maintained for extended periods in antibiotic free media. Regular microscopy, including the high-power examination of hematoxylin and eosin (H&E) stained cells, showed no bacteria, nor did electron microscopic examination of the infected cells. Rather it appears that the incorporation of bacteria-derived sequences was a prior event occurring during or shortly after the stealth adaptation process. The terms “viteria” and “vifungus” were suggested to describe eukaryotic viruses with incorporated bacterial or fungal sequences, respectively.10,29
Of interest are the potential locations at which virus and bacterial sequences would have the opportunity to interact. For intracellular bacteria, such as mycoplasma, borrelia, and brucella, it could be within a dually infected eukaryotic cell. The incorporation of ochrobactrum sequences into a stealth adapted virus probably requires a different explanation. Conventional eukaryotic viruses do not ordinarily grow in bacteria. The genes coding for the growth-restrictive elements may, however, be deleted during the stealth adaptation process. Stealth virus-1 is cytopathic for cells of multiple species, including insects. There is also evidence of stealth adapted viruses affecting bacteria. For example, atypical bacteria were cultured from the feces of the stealth virus-1 infected CFS patient. Moreover, transmissible cytopathic activity was subsequently retrieved from the atypical bacterial colonies (unpublished). Ochrobactrum bacteria are generally regarded as being soil-based, although Ochrobactrum anthropi can occasionally infect humans.30 Humans can, however, be exposed to these bacteria in consumed food 31. These bacteria may also potentially reside in the gut microbiota.
Unlike the non-coding cellular sequences, many of the identified bacteria sequences have open reading frames and would presumably lead to the formation of proteins. The bacteria proteins, including enzymes, may potentially contribute to the virus replicative process and to the induced cytopathology. The proteins could also potentially function as a makeshift capsid for the intercellular transfer of replicated nucleic acids. To do so, the bacteria proteins would presumably need to self-assemble into cage-like structures, which are able to enclose strands of nucleic acids.
Although speculative as to its relevance, it is interesting to relate the incorporation of bacteria sequences to a self-healing process, which occurs during the culturing of stealth adapted viruses. The healing process is associated with the production of self-assembling aliphatic and aromatic chemical compounds. The assembled materials are referred to as alternative cellular energy (ACE) pigments.32 These materials can attract an external force called KELEA (Kinetic Energy Limiting Electrostatic Attraction), which drives the ACE pathway.33–37. Enhancing the ACE pathway can lead to the suppression of the virus induced CPE.32 It would not be surprising, therefore, if some of the bacteria gene-coded proteins were to be subsequently identified as contributing to the formation of ACE pigments. These pigments could potentially prolong the infection process. Bacteria proteins might also be expected to lead to intracellular protein aggregates and to trigger the unfolded protein response.38 This response can also potentially lead to a delay in virus-induced cell death, thereby, allowing for a more prolonged symbiotic relationship. A diverse array of intracellular and extracellular aggregates can be seen by electron microscopy in stealth adapted virus-infected cultured cells and in the biopsied tissues from infected individuals.
The relocation of genetic sequences from their normal cellular or bacteria origin to a virus can be viewed as a passive hijacking of the sequences by the virus or as a loss of the cell’s or the bacteria’s capacity to restrain some of their own genetic sequences from deserting and moving elsewhere. In this context and to help in conveying an understanding of the process, the relocating sequences are being referred to as renegade sequences. In reality, the capturing of cellular sequences by viruses is not a new phenomenon.39 Indeed, even regular SCMV encodes five copies G-protein coding gene, which was presumably originally cellular in origin.40 What is different with regards to the present findings is the concept that the incorporated genes could potentially come to comprise an essentially autonomous, replicating, cytopathic, renegade virus. In other words, once the replicative process involving cellular sequences is underway, there may be no longer a need for remaining sequences of the initiating stealth adapted virus infection. In this regard, stealth virus-1 may be relatively early in its evolution with its retention of extensive SCMV-related sequences.
Viruses are known to engage with noncoding RNA elements within the cell. The interactions can either be advantageous or inhibitory to the growth of the virus, depending on what particular RNA element is being affected.41–46 These previous reports describes the cell and the viruses as separate and discrete, although interactive, entities. A fundamentally different concept is the switching of certain noncoding DNA or RNA elements from primarily belonging to the cell to becoming part of the replicating and transmissible virus genome. Since noncoding sequences have specificity in their activities,47,48 the type or types of incorporated sequences could have a bearing on the cytotoxic or tumorigenic effects of the modified (renegade) virus.
Major substitutions of renegade cellular sequences for the original virus sequences can have important diagnostic implications. On the one hand, the presence of bacteria-derived sequences in stealth adapted viruses can potentially lead to mistaking a virus infection for a bacterial infection. Examples where this error is probably occurring include i) Attributing so called chronic Lyme disease to an active infection by Borrelia burgdorferi.49 Indeed, there is good unpublished evidence from performing stealth virus cultures in many patients diagnosed with chronic Lyme disease, that the individuals are virally infected. It remains to be shown that the viruses contain borrelia-related genetic sequences. ii) Also mistaking Morgellons skin disease for an infection with Borrelia, Agrobacteria, or other parasites.49–51 There have been no examples of actually culturing bacteria from these patients. Many of those tested, however, have had positive stealth virus cultures. iii) Linking CFS with mycoplasma52,53 or with brucella infection. An interesting comment that was made by a mycoplasma researcher was that the mycoplasma sequences he was identifying were actually slightly different from those of the known mycoplasma species. Some CFS patients have been tested for brucella antibodies and found to be positive. iv) Defining PANDAS (Pediatric Autoimmune Neuropsychiatric Disorders Associated with Streptococcal Infections) as a bacteria-induced disease.54, 55 Again, on close analysis there is often a discrepancy between the anti-streptolysin O and anti-DNase B antibody testing in patients diagnosed with PANDAS (personal observation). v) Associating bacteria, such as Porphyromonas gingivalis56,57 or chlamydia58–60 with Alzheimer disease and with rheumatoid arthritis.61,62 The prospect of viteria infections may help in reorienting the thinking about these associations.
An even bigger issue is the many illnesses in which a virus infection is somewhat suspected, yet the regular molecular and serological screenings for known viruses yield negative results. There is typically no accompanying inflammation associated with these illnesses. Prominent examples of these illnesses include CFS, fibromyalgia, autism, multiple sclerosis, amyotrophic lateral sclerosis, bipolar depression, schizophrenia, and Alzheimer disease. Some of the controversies related to viruses causing such illnesses are covered in the references.63–78 Glioblastoma is a further example of a disease with suggestive evidence of an underlying virus infection.79–81 A few cases tested by stealth virus culture did give positive results. Until disproven, many persisting illnesses, including cancers, should be studied for evidence of a possible contribution by an ongoing infection with stealth adapted/renegade viruses.
The brain is particularly susceptible to symptomatic illness due to stealth adapted viruses.76–78 This is because of the spatial distribution of the various aspects of brain functioning. It is unlike other organs in which limited localized cell damage can be easily compensated by overactivity elsewhere in the same organ. There has been an overall increase in mental illnesses and this may well be due to an epidemic of stealth adapted virus infections.
Stealth adapted/renegade viruses can best be detected using virus cultures that take into account the production of ACE pigments. The presence of disease-causing viruses can be further confirmed by inoculation of the cultured material into animals.28 The detection of stealth adapted induced illnesses is important in disease prevention and is relevant to the safety of the nation’s blood supply. It is also relevant to the risk of vaccine triggering of a tissue damaging immune response against some of the normally non-immunogenic components in these viruses. Fortunately, the body’s ACE pathway has the capacity to suppress stealth adapted viruses. Clinical studies are needed to evaluate the effectiveness of the various means of enhancing the ACE pathway in the therapy of stealth/renegade virus infections.82–90
The process of stealth adaptation of viruses has been extended beyond the deletion or mutation of the genes coding for the relatively few components normally targeted by the cellular immune system. Stealth adaptation can also involve the incorporation of added sequences, which can be cellular and/or microbial in origin. The incorporated sequences can lead to the further elimination of the originating virus sequences, with the reformed virus retaining the capacity for replication and transmission and, thereby, to cause diseases. The switching of cellular and/or bacterial sequences into the virus world is likened to the actions of a renegade. The term is, therefore, being introduced as an added description of stealth adapted viruses. Conventional serological and molecular assays for viruses are inadequate as a screening method for renegade viruses. Many chronic illnesses, including some cancer, are likely being caused by renegade viruses. They are best treated by enhancing the alternative cellular energy (ACE) pathway.
I am grateful to the individuals who were unswayed when learning that senior Public Health officials had declared that stealth adapted viruses do not exist and that my clinical testing for the viruses had put the Nation’s health in Immediate Jeopardy.
None.

©2019 Martin. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.