Journal of
eISSN: 2373-6453


Research Article Volume 5 Issue 2
0#Both authors have equally contributed to this study
1Department of Medical Microbiology, Jomo Kenyatta University of Agriculture and Technology, Kenya
2Center for Virus Research, Kenya Medical Research Institute, Kenya
3Diagnostic virology department, Max von Pettenkofer-Institute (MVPI), Germany
Correspondence: Eddy Okoth Odari, Department of Medical Microbiology, Jomo Kenyatta University of Agriculture and Technology P.O. Box 62000-00200. Nairobi, Kenya
Received: February 03, 2017 | Published: February 10, 2017
Citation: Omolo GA, Nitschko H, Lwembe R, Maiyo AK, Odari EO (2017) Evaluation of A Reverse Transcriptase (Rt) Loop Mediated Isothermal Amplification Assay For Detection Hepatitis C Genotypes 1-4 Viruses Under Limited Logistical Conditions. J Hum Virol Retrovirol 5(2): 00145. DOI: 10.15406/jhvrv.2017.05.00145
The loop-amplification mediated isothermal amplification (LAMP) presents characteristics that overcome the limitations associated with the current nucleic acid based technologies (NATs), hindering their use in the low resource settings, hence overreliance on serological assays which miss hepatitis C virus (HCV) during the long seroconversion period. The LAMP assay would be ideal for early detection of HCV in routine diagnostics and blood transfusion setups in such settings, drastically reducing transmission related transfusion. This study validated and tested a reverse transcriptase-LAMP for HCV detection under limited logistical conditions in a low resource setting.
Under stringent laboratory conditions, analytical sensitivity and reproducibility were performed using a panel of HCV positive plasma of genotypes 1a, 1b, mixed 1a/1b, 2b, 3a and 4. Cell culture supernatants of HIV-1 B and plasma samples for Hepatitis B virus were used for specificity testing. Upto 227 plasma including 70 (40 RNA positive and 30 negative) from German patients and 157 (43 RNA positive and 114 negative) from Kenyan patients were tested. Kenyan samples were obtained from 121 sero-positive plasma screened from 1121 participants of various cohorts in Kenya.
Although LAMP detected upto 102 IU/mL for genotypes 1a,1b and 2b, a lower detection threshold was established at 103 IU/mL. Overall sensitivity was 94% (PPV 98%) and specificity was 98% (NPV 96%) for RT-LAMP. Sub optimal detection was noted for genotypes 2b, 3a and 4. Sequence analysis revealed mismatches affecting stringency of primer binding at the F1, B1 and the LB primer targets.
RT-LAMP shows potential for early HCV diagnosis and screening in low resource settings. Its robustness is however genotype dependent and can be enhanced by designing primers targeting circulating and suspected genotypes. More studies should be done on the possibility of designing multiplex RT-LAMP primers to capture a wide variety of genotypes. The assay remains simple, rapid, and cost effective for nucleic acid detection and is ideal for use in the limited resource settings.
Keywords:RT-LAMP, Hepatitis C virus (HCV), Kenya, Limited logistical condition
LAMP, Loop-Amplification Mediated Isothermal Amplification; NATs, Nucleic Acid Based Technologies; HCV, Hepatitis C Virus; HNB, Hydroxynaphthol Blue; IDUs, Injecting Drug Users; cDNA, Complementary DNA; dNTP, Desoxynucleotide Triphosphates; DRC, Democratic Republic of Congo
Hepatitis C Virus remains a major public health concern in many parts of the world due to its long sero-conversion period presenting it as a high risk in transfusion related transmission. The transfusion related transmission still remains a concern in many developing countries and resource related settings1 which are highly dependent on serological tests for diagnosis and routine screening of blood meant for transfusion. Although 15-20% of the infected clear the virus spontaneously, 80-85% of those infected are thought to progress to chronic liver infections characterized by liver cirrhosis and hepatocellular carcinoma.2 Genetic diversity of HCV has been thought to influence the treatment outcomes3 and six genotypes denoted as genotypes 1-6 are currently known.4 The geographical distribution of these genotypes show that genotypes 1 - 3 circulate mainly in the United States of America (U.S.A) and Europe, genotypes 4 and 5 are found in some parts of Africa mainly in Egypt and the Democratic Republic of Congo (DRC) for genotype 4 and South Africa for genotype 5 with genotype 6 so far reported only in Hongkong.4 Genotypes specifically associated with Africa are determined as genotype 1a, 2b and 4, all of which have been reported in Kenya.5,6
The long window period associated with the HCV virus is a major reason for its poor diagnosis in many laboratories in the developing world and the low resource settings. Many laboratories in the latter regions mainly rely on detection of antigen or antibodies to the virus.7-9 A number of these serological assays have even been designed as combination assays intended to reduce the window periods.10,11 However, some studies have demonstrated the inefficiency of the same combination assays to detect two proteins with a similar degree of sensitivity in a single run12 as claimed by the manufacturers. The gap occasioned by the long seroconversion period leaves only the Nucleic Acid based Tests (NATs) as reliable tools for HCV diagnosis early within the window period. The limitations of the conventional and widely used nucleic acid based technique (NATs) of high costs, long time to results, sophistication, need for specialization among others have been well elucidated.13 Since its development about 15 years ago,14 loop-mediated isothermal amplification (LAMP) has presented characteristics that are ideal as an alternative for use in the limited resource settings. The characteristics of simplicity, cost-effectiveness and rapidity have attracted the assay for evaluation of LAMP for detection of a number of pathogenic microorganisms. The LAMP assay uses a set of primers targeting six arbitrarily designated regions F1, B1, FIP, BIP, LF and LB under isothermal conditions (normally a temperature between 60-65°C). The inclusion of Bacillus stearothermophilus (Bst) DNA Polymerase with strand displacement capability ensures loop formation which become the targets of amplification resulting in the generation of high amounts of magnesium pyrophosphate (Mg2O7P2) deposits. These Mg2O7P2 deposits then become the targets of detection either visually by use of a color detection dye, or by turbidimetric reading or detection in real time by use of a turbidimeter.15
With addition of a reverse transcriptase enzyme RT-LAMP has been evaluated for a number of viruses including for Hepatitis C virus detection.16-18 However, these studies have been carried out under stringent laboratory conditions with clinical samples mainly from the Asian continent. No study has so far been reported on the efficacy of HCV RT-LAMP under limited logistical conditions in sub-Saharan Africa. This study therefore aimed at evaluating the RT-LAMP in detecting HCV under stringent in Europe (Germany) and comparing the results with those performed under limited logistical conditions in sub-Saharan Africa (Kenya).
Comparative assays
Abbott m2000rt system (Abbott Laboratories, North Chicago, IL, USA) served as a comparative assay for samples analyzed in Germany, whereas multiplex RT-PCR was the only available comparative assay for Kenyan samples tested in Kenya. The Abbott m2000rt system is a commercially available assay used to detect and measure the amount of HCV nucleic acid particles in real time using fluorescent labeled oligonucleotide probes. The fluorescent signals are always proportional to the log of the virus RNA measured in International Units per milliliter (IU/mL).
Nucleic acid extraction and LAMP amplification
Reagents/equipment: Roche MagNAPure (Roche Diagnostics, Mannheim, Germany) and High Pure Viral Nucleic Acid (cat. no. 11858874001; Roche Diagnostics, Unterhaching, Germany) kits were used for extraction of viral nucleic acid from the plasma samples. The MAST IsoplexTM RNA Amplification kit (Mast Diagnostica, Reinfeld, Germany) was used for LAMP assay amplification. The RNA amplification kit contained 8 U/µL Bst polymerase enzyme, 20 U/µL reverse transcriptase enzyme (Roche Transcriptor reverse transcriptase), water, detection dyes (V13 indicator dye for real time turbidimetric detection and hydroxynaphthol blue (HNB) for visual color detection) and an already constituted 2×reaction mix (RM). The reaction mix contained dNTPs (2.8 mM), KCl (20 mM), (NH4)2SO4 (20 mM), Tris buffer (40 mM, pH 8.8), betaine (1600 mM) and MgSO4 (16 mM). For real time turbidimetric readings the Loop Amp real-time turbidimeter (LA-200m; Teramecs, Kyoto, Japan) was used.
Reaction primers
All the primers used targeted the 5’ UTR conserved gene (Accession number: GQ418245). Whereas the LAMP primers used had previously been described,16 in-house primers with a few modifications to include wobble bases were generated and used during the RT-PCR amplification. These primers for RT PCR were designated as outer primers; HCVN 01– GGC GAC ACT CCA CCA TRR A (forward), HCVN 02 – GTG CAC GGT CTA CGA GAC C (reverse), HCVN 08 –TAC TCA CCG GTT CCG CAG A (Reverse), Inner primers HCVN 03 – CAC TCC CCT GTG AGG AAC T – 3’ (forward), HCVN 04 - CCC GGG GCA CTC GCA AGC A (Reverse) with HCVN 02 and HCVN 08 being used during reverse transcription to generate HCV cDNA. All primers were synthesized and supplied by Ella Biotech (Ella Biotech GmbH, Munich - Germany).'
Hepatitis C samples
Validation panel:An in-house sample panel of known HCV positive plasma were obtained from the Max von Pettenkofer Institute in Munich-Germany. The plasma samples with HCV viral loads V1529272 (1.4 x 106 IU/mL), V1529317 (2.3 x 106 IU/mL), V15103800 (6.1 x 106 IU/mL), V0728602 (2.1 x 106 IU/mL), V1529319 (5.5 x 106 IU/mL), V0743586 (1.6 x 106 IU/mL) representing genotypes 1a, 1b, mixed 1a/1b, 2b, 3a and 4 respectively were used during the validation testing of the RT- LAMP.
Clinical samples from patients: A total of 227 plasma samples including 70 (40 RNA positive and 30 negative) from German patients and 157 (43 RNA positive and 114 negative) from Kenyan patients were tested. The Kenyan RNA positive plasmas were part of the 121 sero-positive samples screened from a total of 1121 persons of various cohorts including injecting drug users (IDUs), female sex workers, blood donors and patients attending various outpatient clinics in Nairobi Kenya.
Ethical clearance: For Kenyan samples, ethical clearance was obtained from the Scientific and Ethical Review Unit (SERU) of the Kenya Medical Research Institute (KEMRI) in Nairobi (protocol number KEMRI/SERU/CVR/008/3179), while the German samples were used with permission from the routine diagnostic laboratory of the Max von Pettenkofer-Institute (MVP) in Munich.
Generation of quantitation panel for correlation analysis and test control samples
Total nucleic acid was extracted from plasma sample panels of genotypes 1a, 1b, mixed 1a/1b, 2b, 3a and 4 using an automated system, Roche MagNAPure (Roche Diagnostics, Mannheim, Germany), according to manufacturer’s instructions. Tenfold dilutions were made from the extracted materials and used to constitute quantitation panels for correlation analysis. The dilutions were further maintained as positive control samples during the testing of clinical samples. Negative controls included known HIV-1 genotype B (MVP 899-87) cell culture supernatant, ultra-pure water (H2O), 0.9% sodium chloride (NaCl) and poly (A) from High Pure Viral Nucleic Acid.
Processing of samples by Abbott Real Time
The sample panel as well as clinical samples tested at the Max von Pettenkofer institute were first amplified and quantified by Abbott Real Time assay (http://www.abbottmolecular.com/ static/cms workspace/pdfs/US/51-602146R6.pdf as described by the manufacturer (Abbott Laboratories, North Chicago, IL, USA). The starting sample volume for automatic nucleic acid extraction was 1 mL and where the appropriate volume could not be attained, samples were filled up with 0.9% NaCl to 1 mL and then the material processed.
Testing samples by RT-PCR
Reverse transcription: Purified RNA extracts were first reverse transcribed to generate complementary DNA (cDNA) using reverse primers HCVN 02 and HCVN 08. Briefly 10 µM of each reverse primer, 4 µL of 5x buffer (100 µM Tris-HCL, 250 µM KCL, pH 8.4), 2 µL DTT (0.1 M), 2 µL desoxynucleotide triphosphates (dNTP) (5 mM) and 40U reverse transcriptase enzyme (cat. no. 18064-014, Life Technologies, Germany) were constituted for a 10 µL reaction mix. The reaction mix was incubated with 10 µL of the of the extracted RNA template at 45ºC for one hour.
Nested PCR
The PCR reaction was performed in a total volume of 50 µL (for first PCR amplification) containing 31.7 µL PCR water, 5 µL reaction buffer, 2 µL of dNTP, 3µL of each of the primers HCVN 01 and HCVN 02, 0.3 µL of taq Polymerase and 5 µL of cDNA to amplify a 296-bp fragment. In the second round of PCR 3 µL of first round PCR product was added to 47 µL of the PCR mixture similar to the first but with the inner primers HCVN 03 and HCVN 04 for 30 cycles in order to amplify a 253-bp fragment as visualized on a 2% agarose gel.
Processing of samples for testing by RT-LAMP assay
Extraction of RNA from plasma:Viral RNA was extracted using High Pure Viral Nucleic Acid Kit (cat. no. 11858874001; Roche Diagnostics, Unterhaching, Germany) according to the manufacturer’s instructions. In this method 200µL of each sample were used as the starting volume for the extraction process and the RNA was eluted in 50µL of elution buffer supplied in the extraction kit (https://cssportal.roche.com/LFR PublicDocs/ras/11858874001 en 16.pdf).
RT-LAMP amplification: The master mix for RT-LAMP reaction was composed of 40 pmol for each inner primer (FIP/BIP), 5 pmol for each outer primer (F3/B3), 25 pmol for each loop primer (LF/LB) primers. Further 12.5 µL of the reaction mix, 1µL of Bst polymerase enzyme, 0.5 µL reverse transcriptase, 1µL of the detection dye were added and topped up with water for a volume of 20 µL. A total of 5 µL of the RNA template was added to the 20 µL master mix so as to make a total reaction volume of 25 µL and the reaction mix incubated at 63ᵒC for 60 min in a LoopAmp real-time turbidimeter (LA-200 m; Teramecs, Kyoto, Japan). The threshold value (Abs) equivalent of an optical density above 0.1 by the turbidimeter indicated a positive detection.
Nucleic acid sequencing
The process of sequencing involved 3 phases which were performed according to the manufacturer’s protocol using a Beckman Coulter CEQ 8800 system (Agencourt Bioscience Corporation, Beverly, MA, United States). This sequencing method is based on use of didesoxynucleotides.19 The sequence results generated were edited and finally aligned using a Bioedit software20 (Ibis Biosciences-Carlsbad, CA, USA) and blasted in the HCV database to identify the genotypes.
Testing analytical parameters for RT-LAMP
Analytical sensitivity and the lower limit of detection (LOD): Analytical sensitivity is the minimum number of copies in a sample which can be measured accurately by an assay.21 It is currently recommended that lower limit of detection be determined as the end point dilution at which 50% of the tested samples are positive.22
The tenfold dilution from the validation panels were made ranging from 106 IU/mL to 101 IU/mL and each tested ten times and results recorded as positive or negative for each viral load concentration by the LAMP assay and the percentage of positive detections calculated.
Analytical specificity: Analytical specificity was tested using RNA templates HIV-1, Hepatitis B virus DNA with virus concentrations between 103 to 106 copies/mL.
Precision
Inter-assay precision was evaluated by testing the concentrations of the panel on 3 different days and measuring the time to detection (Tt) values using a real time turbidimeter. The coefficient of variation (CV) was thus determined based on the Tt values. Since the testing was not aimed at testing the real time turbidimeter, intra-assay testing was omitted.
Statistical analysis
The sensitivity, specificity and predictive values for HCV RT-LAMP were calculated for clinical samples based on a confidence interval of 95%. Spearman’s correlation test was used to compare viral load concentrations against the time to detection using SPSS version 16.
Analytical validation
The RT-LAMP amplification was determined by typical ladder-like banding patterns (Figure 1a) and the visual detection showed clear distinction of positive results using hydroxy naphthalol blue (HNB) dye (Figure 1b).
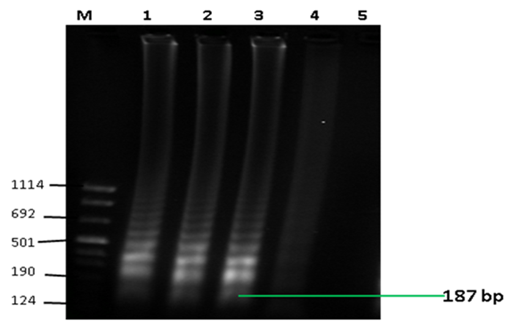
Figure 1a Detection of LAMP products by gel electrophoresis. The figure shows typical ladder-like banding patterns associated by LAMP assay expected at 187 bp. Lanes M: molecular weight marker VIII (Sigma-Aldrich, LLC), 1-3: positive results, 4 : unspecific result, 5 : negative result.

Figure 1b Visual detection of LAMP products. The figure shows positive and negative results by use of hydroxy naphthol blue (HNB) dye. Tubes 1 -3 show positive results showing blue colour change while tube 4 shows a negative result by a clear purple colour change.
Analytical sensitivity: Analytical sensitivity of RT-LAMP assay showed a lower detection threshold at a viral load of 103 for genotype 1a, 1b and 2b where the rate of detection was above 50% (Figure 2). Viral loads of 102 were detected at about 40% rate with the assay detecting down to 51 IU/mL albeit only at 20% detection rate for genotype 1a and 1b. The sensitivity for genotypes 3a and 4 were relatively low with a potential lower limit of detection only for samples above 104 IU/mL. Detection rates of samples below 103 IU/mL were less than 30% or none for genotypes 3a and 4. The LAMP assay did not detect all the 30 HCV negative samples, including those that had been determined positive for HIV and Hepatitis B infections.
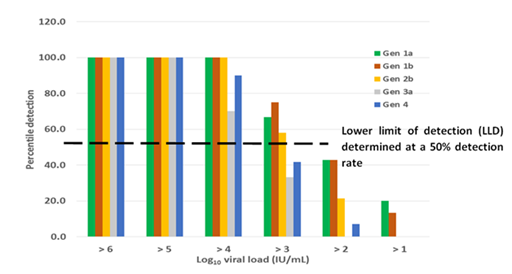
Figure 2 Detection rate of LAMP on selected genotypes. A higher detection rate is shown among genotypes 1a, 1b and 2b with a lower limit of detection potentially shown at viral loads within 103 IU/mL. There is a potential suboptimal detection of LAMP for genotypes 3a and 4.
Reproducibility: Reproducibility of the assay was analyzed by testing the precision based on the time taken to threshold detection using a real time turbidimeter. A high precision was observed for all samples tested for samples above 103 IU/mL for all the genotypes (although genotype 1b showed a higher variation at viral load 103 IU/mL) tested except for genotype 3a which recorded a poor precision for viral loads of 104 and 103 IU/mL (Table 1) where variations above 10% were observed.
Genotype |
Log10 viral load (iu/ml) |
Time to threshold detection in minutes |
CV |
|||
Test D1 |
Test D2 |
Test D3 |
Mean detection time |
|||
Gen 1a |
6 |
24.12 |
24.18 |
25.11 |
24.28 |
0.02 |
5 |
29.36 |
31.54 |
30.54 |
30.29 |
0.04 |
|
4 |
34.24 |
34.06 |
36.24 |
34.51 |
0.03 |
|
3 |
39.06 |
44.15 |
44.14 |
42.27 |
0.07 |
|
2 |
42 |
48.13 |
47.34 |
45.49 |
0.07 |
|
1 |
- |
- |
- |
- |
- |
|
Gen 1b |
6 |
23.42 |
23.24 |
23.06 |
23.14 |
0.01 |
5 |
26.36 |
26.54 |
24.54 |
25.49 |
0.04 |
|
4 |
28.33 |
28.06 |
29 |
28.28 |
0.02 |
|
3 |
46.18 |
37.54 |
31.48 |
38.24 |
0.19 |
|
2 |
60 |
48.02 |
56.06 |
54.41 |
0.11 |
|
1 |
- |
56.21 |
- |
- |
- |
|
Gen 1a/1b |
6 |
N/A |
N/A |
N/A |
N/A |
*** |
5 |
27.36 |
26.12 |
27.01 |
26.5 |
0.02 |
|
4 |
31.08 |
30.12 |
30.08 |
30.26 |
0.02 |
|
3 |
35.12 |
34.54 |
37.16 |
35.37 |
0.04 |
|
2 |
47.13 |
51.34 |
52.28 |
52.15 |
0.05 |
|
1 |
- |
58.23 |
- |
- |
||
Gen 2b |
6 |
N/A |
N/A |
N/A |
N/A |
*** |
5 |
33.52 |
34.48 |
34.59 |
34.12 |
0.02 |
|
4 |
34.14 |
37.11 |
40.13 |
37.08 |
0.08 |
|
3 |
47.02 |
55.36 |
53.3 |
51.53 |
0.08 |
|
2 |
59.56 |
- |
- |
- |
||
1 |
- |
- |
- |
- |
||
Gen 3a |
6 |
24.06 |
23.24 |
N/A |
23.39 |
*** |
5 |
25.18 |
24 |
N/A |
24.35 |
*** |
|
4 |
33.18 |
30 |
39 |
34.04 |
0.13 |
|
3 |
40 |
47.13 |
51.34 |
46.1 |
0.12 |
|
2 |
- |
- |
- |
- |
- |
|
1 |
- |
- |
- |
- |
- |
|
Gen 4 |
6 |
34.12 |
35.13 |
34.36 |
34.32 |
0.02 |
5 |
36.23 |
39.04 |
37.28 |
37.31 |
0.04 |
|
4 |
48.46 |
47.24 |
55.31 |
50.2 |
0.09 |
|
3 |
60 |
- |
- |
- |
- |
|
2 |
- |
- |
- |
- |
- |
|
1 |
- |
- |
- |
- |
- |
|
Table 1 Inter-Assay Precision test for LAMP assay on a panel of known HCV viral loads
***Co-efficient of variation (CV) not calculated due to inadequate number of tests done or lack of time to threshold values.
N/A: Tests not done due to lack of or inadequate volumes of samples
Dashes depict negative results
Clinical validation
A total of 227 plasma samples were tested on RT-LAMP assay where the overall sensitivity of 94% (PPV = 97.5%) and specificity of 98.6% (NPV = 96.6%) were determined (Table 2). On the various groups of samples, a lower sensitivity was observed for German samples mainly as a result of the 5 samples (4 for genotype 3a and 1 for genotype 1b) which could not be detected. For Kenyan samples, a reduced positive predictive value was due to 2 samples that were determined as false positives by RT-LAMP. It was also noted that although all Kenyan were detected, genotype 2b and 4 were detected late after 40 minutes as opposed to the genotype 1a which were detected in less than 35 minutes.
Sample Group |
Number of Samples |
Parameter Tested |
|||
Sensitivity (n/N) (%) |
Specificity (n/N) (%) |
PPV (%) |
NPV (%) |
||
Overall |
n = 227 |
(78/83) (94%) |
(142/144) (98.6%) |
97.5 |
96.6 |
German |
n = 70 |
(35/40) (87.5%) |
100 |
100 |
100 |
Kenyan |
n = 157 |
(43/43) (100) |
(112/114) (98.2) |
95.6 |
100 |
Table 2 Sensitivity and specificity analysis of RT-LAMP assay on various groups of samples (n = 227)
PPV: Positive predictive value
NPV: Negative predictive value
Sub optimal detection of genotype 3a and 4
A parallel shift was however shown with genotype 3a and genotype 4 indicating a sub-optimal amplification for the genotypes, a fact which could be potentially true for genotype 2b which also showed a late detection threshold time for all the sample dilution panels (Figure 3). It is thus not clear from the results whether the false negative results recorded here as a result of non-binding of the primers to the viral RNA or the need for a longer run time for LAMP. Sequence analysis of the various genotypes showed mismatches at the F1 the B1 and the LF primer target sites mainly for genotypes 2b, 3a and partially 4 (Figure 4).
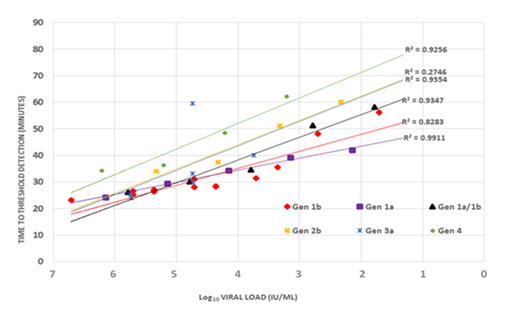
Figure 3 A correlation analysis between the viral load and the time to detection. An inverse correlation observed for all genotypes. Genotypes 2b, 3a and 4 showing a parallel shift indicative of sub-optimal amplification of these samples by the LAMP assay.
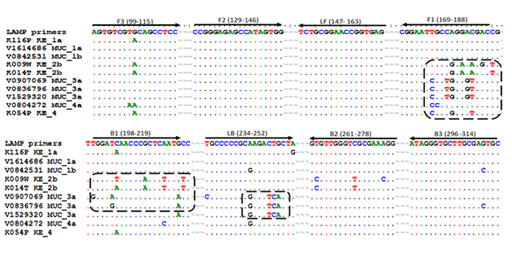
Figure 4 Mismatches at specific LAMP primer targets shown by a selected group of samples showing sub-optimal amplification from Germany and Kenya. An increased number of mismatches (in rectangular dotted rings) showed for HCV genotypes 2b, 3a and 4 at positions F1, B1, and LB. The highest number of mismatches was observed for genotype 3a (13 mismatches) 2b (12 mismatches) and 4 (5 mismatches). Genotypes 1a and 1b which showed optimal performance had mismatches less than 5.
Effective diagnosis of HCV within the sero-conversion period remains a major challenge in transfusion medicine and especially in the low resource countries where serological assays remain the major diagnostic systems for use. The long amount of time, sophistication and the high costs associated with the Nucleic Acid based Techniques (NATs) hinder their use in routine diagnosis and screening of blood in these low resource settings. This study reveals HCV RT-LAMP as a reliable alternative to the available NATs for diagnosis of hepatitis C virus in plasma and compares the performance of the same assay platform for selected genotypes. Although a study17 has evaluated LAMP on genotypes 1-6, a number of studies published16,18 have dwelt mainly on genotypes circulating in Asian countries and the Arab world. This study presents the success of RT-LAMP assay under limited logistical conditions and further shows the differences in amplification of LAMP on a selected genotypes 1a, 1b, mixed 1a/1b, 2b, 3a and 4. The results obtained by the clinical samples both of Europe and sub-Saharan Africa and the success in their detection is a clear demonstration of robustness of HCV RT-LAMP under both favorable (stringent) and limited logistical conditions. Studies have shown that HCV RNA become detectable in sera between 7 – 14 days after exposure, before the detection of aminotransferase and the antibodies - all coming between 4 to 10 weeks.23 The sensitivity by RT-LAMP in this study therefore demonstrate that its use show a potential to reduce the window period for HCV detection to only a maximum of 14 days, similar to any available NAT. In fact some studies have even shown that HCV RT-LAMP is more sensitive compared to nested RT PCR and of similar sensitivity to real – time RT PCR.18 Of the 121 seropositive Kenyan samples, only 43 were detected by HCV nested RT-PCR whereas 45 were detected by RT-LAMP (2 samples noted as false positive), an indication that the samples not detected may have been as a result of no or low level of HCV RNA normally as a result of spontaneous clearance of the viruses. It can however not be concluded whether the 2 aforementioned samples noted as false positive by RT- LAMP were truly false positive, since other studies have reported a higher sensitivity by RT- LAMP compared to nested RT PCR.18 Failure to test the Kenyan samples with both real time (q) PCR (since real time RTPCR was not available for testing the Kenyan samples during the course of this study) as well as nested PCR, especially for the two false positive samples was therefore a major limitation of this study.
The lower detection threshold established at approximately 103 IU/mL in this study corresponds to other studies17 which have demonstrated a detection rate of 80% for samples of 50 IU/rxn (approximately 2200 IU/mL) with 10 IU/rxn (400 IU/mL) only detected at 40% rate. The findings of the aforementioned study as well as the results of this study are also not significantly different from the lower limit detection of 660 IU/mL determined some further studies.18 The lower detection limits determined are not only important in detecting the virus early enough, but could also be important in designing HCV RT-LAMP as a quantitative assay for monitoring the success in HCV treatment which has been established to be genotype specific.3 Indeed, some studies done on HIV have shown RT-LAMP to be an effective tool in semi-quantitation of viral load as an avenue for effective monitoring of treatment success,12 a fact that needs to be evaluated for HCV as well. Monitoring of success in HCV treatment through viral load measurement is not only important due to toxicity of the drugs to the liver and the genotype specificity of the drugs but also due to the fact that the current drugs for HCV treatment are very expensive and beyond reach of many, especially the patients living in the low resource countries.
The specificity shown in this study depict this assay as a reliable tool for HCV specific detection especially in the tropical countries like Kenya where HIV, HBV and other infections like malaria are of high prevalence. The RT-LAMP assay from this study and other studies17 can therefore be presented as devoid of cross-reactivity with other viruses and parasites such as malaria parasite. It is however important to note that although this study and others mentioned for HCV have not reported cross-reactivity, other studies done with RT-LAMP12,24 have indicated a potential for non-specific binding leading to false positive results. In their work Odari and the group12 suggests that the complex reaction process coupled by the number of primers used for any LAMP reaction are a potential source of unspecific binding leading to the generation of Magnesium pyrophosphate (Mg3(PO4)2) deposits generated by detectable by the turbidimetric reading. If the latter assertion is true, then RT-LAMP for HCV detection would not be an exception hence the need to improve the assay for elimination of potential false positive results. False positive results unnecessary treatment leading pressure on the existing HCV treatment regimen.
The ability to use a simple and cost effective hydroxynaphthol blue (HNB) dye for visual detection relieves this assay of the complex post amplification process of gel electrophoresis. This post amplification process has been established to be a major cause of contamination in PCR related work. In deed the relative stability shown for LAMP products25 has been thought to be a major source of carry over contamination12 since these products are not easily degradable. In the absence of other intercalating dyes and turbidimetric reading, the HNB dye would be ideal for use in the low resource countries as this will eliminate laboratory contamination as well as reduce the time and cost due to a simple negative or positive result as a result of a simple colour change. The use of colour dyes would also eliminate the need for highly trained laboratory staff, thus could be used for blood screening at the established blood collection sites before transportation to the regional or national blood banks.
The parallel shifts shown with genotypes 3a and 4 in this study are a result of potential late detection of indicating sub-optimal detection of these samples with the primers used. Further analyses with the primer sequences showed mismatches at established positions for the sub-optimally detected genotypes including genotype 2b thus suggesting primer specificity for LAMP assay. Although the issue of mismatches and their effect on amplification or quantification is not unique for LAMP assay;26-31 it erodes the major advantages that LAMP has portrayed over other NATs. Therefore, designing LAMP primers remain a major focus in the sensitivity of this assay platform. Although Nyan and Swinson17 demonstrated that a universal primer could detect HCV genotypes 1 to 6, it suffices to note that the efficacy of such primers is reduced with the number of mismatches as can be observed in Figures 3 and 4 in this study. Therefore, knowledge of circulating genotypes is necessary in the establishment of RT-LAMP in detection or screening of HCV. The best strategy would be to develop a mixture of primers targeting local and suspected genotypes so as not to miss any imported strains. The efficacy of such primers however would need to be evaluated further.
In conclusion, we note that the rapidity, sensitivity and specificity coupled with its simplicity present HCV RT-LAMP is as a suitable assay for diagnosis of acute infection and screening for HCV infection in donor blood in low resource settings. The success of the latter however would largely depend on the type of the LAMP primers used, hence calling for knowledge of circulating genotypes and those genotypes that could potentially be imported as this knowledge would reduce the risk of false negative results. Further measures such as elimination of post amplification procedures for gel electrophoresis should be ensured and visual or turbidimetric reading be employed to ensure true positive results. There is thus the need for further research on the potential use of mixture of primers targeting known HCV genotypes and the use of RT-LAMP as a semi-quantitative tool for HCV viral load measurement in HCV patients undergoing treatment.
We acknowledge the Max von Pettenkofer – institute for providing the sample panels and clinical samples for the patients in Germany and undertaking to genotype a selected number of Kenyan samples, Ella biotech in Munich – Germany for donating primers, the Kenya Medical Research Institute for providing the laboratory space in Kenya, the Kenya National Blood Transfusion center for providing the blood bank samples, the KANCO and ARMUT drop in centers in Watamu and Malindi, Kenya for facilitating clinical sample collection and the department of Medical Microbiology and School of Biomedical Sciences of JKUAT for facilitating this research. Finally we acknowledge the Jomo Kenyatta University of Agriculture and Technology, Research Production and Extension (RPE) and the Kenya National Research Council (NACOSTE) for the financial support.
None.

©2017 Omolo, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.