Journal of
eISSN: 2373-6453


Research Article Volume 4 Issue 2
1French National Reference Center for Arboviruses, French Army Biomedical Research Institute (IRBA), Marseille, France
2Institut Pasteur Paris, France
3CHR Felix Guyon, Saint Denis de la Reunion, France
4Institut Pasteur de Cote d’Ivoire, Cote d’ivoire
5Centre de Recherche pour le Developpement, Universite Alassane Ouattara de Bouake, Cote divoire
6Institut Pasteur du Laos, Laos
Correspondence: Marianne Maquart, Institut de recherche Biomedicale des Armees, Hopital Laveran, 34 boulevard Laveran, 13013 Marseille, France
Received: October 17, 2016 | Published: December 1, 2016
Citation: Monteil VM, Maquart M, Caro V, Jaffar-Bandjee MC, Dosso M, et al. (2016) Circulation of Dengue Virus Type 3 Genotype III in Africa Since 2008. J Hum Virol Retrovirol 4(2): 00128. DOI: 10.15406/jhvrv.2016.04.00128
Dengue virus type 3 (DENV-3) genotype III circulated across East Africa between 1984 and 1993, but no additional case was detected until the return of a European traveler from Cameroon in 2006. Since then, DENV-3 genotype III has been reported in many countries like Tanzania, Senegal and Cote d’Ivoire. Fifteen strains of DENV-3 were isolated from Africa by the laboratory of French National Reference Center for arboviruses between 2008 and 2012. The objective of the present study was to establish the phylogeny of new strains of DENV-3 genotype III isolated in Africa to better understand their origin(s) and diversity. Analyses showed that the Cote d’Ivoire 2008 strain was phylogenetically linked with the strains responsible for all the epidemics that occurred since then in West Africa including Cape Verde. This strain was also phylogenetically linked with the strain isolated in China in 2009. The Tanzanian 2010 strains cluster included the strains isolated in the Union of Comoros, Madagascar and China. Strains isolated from sporadic cases in La Reunion in 2012 clustered with the Thailand strain. These results evidenced the diversity of DENV-3 genotype III isolated in Africa between 2008 and 2012. The impact of air travel, tourism and trades should play a major role in the DENV spreading over continental Africa and African archipelagoes.
Keywords: Dengue virus serotype 3, Genotype III, Dengue virus, Africa, Circulation, DENV-3, DHF, Infections, RNA virus
DENV-3, Dengue Virus Type 3; DHF, Dengue Hemorrhagic Fever; FRNC, French National Reference Center; DENV, Dengue Virus
Dengue virus (DENV) is a mosquito-borne Flavivirus, which is an enveloped single-stranded positive RNA virus. Most dengue virus infections are asymptomatic, with an estimated 390 million infections per year occurring across approximately 128 endemic countries, of which 96 million manifests clinically.1
Emergence of the disease is actually associated to the large expansion of its vectors (Aedes species) particularly by the trade of used tires that are thought to explain the dispersal of eggs and immature forms of these mosquitoes into new territories.2 Moreover, traveling within endemic areas, climate change favoring a rapid adaptation of the insects into new geographic locations, and lack of medical surveillance have greatly contributed to the increase in dengue infections worldwide.3
The virus is endemic in tropical and sub-tropical areas, mainly in Asia and South and Central America, but reports of dengue circulation are increasing in Africa. Four antigenically distinct serotypes (DENV1-4) circulate simultaneously in endemic countries, with 60-70% identity in nucleotide sequences4 and approximately 70%5 in deduced amino acid sequence identity among the four serotypes. Secondary disease with 3 DENV-2 and -3 are more likely to result in DHF as with DENV-1 and-4. Nevertheless, other factors besides serotype may play a role in disease severity.6 Dengue virus infections in Africa are low compared to South-East Asia and the Americas. This distribution is due to two main factors: worldwide dengue virus activity and the popularity of certain countries as tourist destinations.7 Effectively, epidemics may remain undetected until diagnosis is assessed in another country from a returning infected visitor. Moreover, travelers may be more susceptible to dengue infection than locals, particularly travelers from non-endemic to endemic areas.8 Dengue is also under recognized in Africa because of low awareness by health care providers, other prevalent febrile illnesses, and lack of diagnostic testing and systematic surveillance.9 Dengue epidemics have been reported in Africa since the 19th century, in countries including Zanzibar, Burkina Faso, Egypt, South Africa, and Senegal. Available data suggest that dengue is endemic to 34 countries across all regions of Africa. Dengue virus is still expanding worldwide to new territories with the first epidemic in Cape Verde in 2009 with more than 17,000 cases.7 It is likely that all four subtypes of the dengue virus are present but the lack of formal laboratory testing or surveillance initiatives means that it is difficult to verify this.8
Epidemiological studies have shown that some strains of dengue might be associated with increased severity than others. The serotype swing in some regions caused enhanced severity of dengue outbreaks.10 In this context, surveillance of more virulent strains is now considered essential public health activity. DENV serotype 3 (DENV-3) was first isolated during a DHF epidemic in Manila in 195611 and is now circulating in Asia and South America.12,13,14 This serotype is often associated with more severe diseases and dengue hemorrhagic fever than other serotypes.15,16,17 Little is known about DENV-3 activity in Africa. Serotype 3 was first detected in Africa during a 1984 epidemic event in Pemba (Mozambique), where at least 45% of the local population was affected.18 This high proportion could be explained at least in part by the naïve immune status of the population against this new serotype. Since then, sporadic occurrences of the serotype 3 were reported, such as in Kenya in 1991 and in Somalia in 1993.19 DENV-3 was responsible for 2 epidemics in Sudan in 2004-2005 and 2010.20 The latter affected over 3765 people.21 In West Africa, DENV- 3 circulation was first published in 2008 in Cote d’Ivoire, during a yellow fever alert.22,23
Moreover, subtle changes such as the ones at the lineage level could alter the potential of the dengue virus to cause severity, as it has already been observed in Sri Lankan DENV-3 strains.24,25 The E protein is the major target gene for genetic variations with a possible impact on virulence.26,27 Nucleotidic comparison of the envelope (E) gene sequences led to the identification of 5 genotypes within serotype 3.17,28 Particularly, genotype III is now composed of strains from Samoa, Sri-Lanka, India, Africa, and America since 1994. DENV-3 genotype III (Sri Lanka/India) was identified in Brazil during the epidemics of 2001 to 2004, and caused one of most severe epidemic with the largest number of reported cases, greatest severity of clinical manifestations in primary infections in particular, and the largest number of confirmed deaths.29 DENV-3 genotype III was recently considered as an emerging DENV genotype in Southeast Asian countries but also in Africa. The close relatedness between strains isolated in Mozambique in 1985 and in India strongly suggested that DENV-3 Genotype III could have been imported in Africa from India17 and led to the establishment of a stable East African group from Mozambique, Kenya, and Somalia isolated from 1985 to 1993.30 In 2008, the Institut Pasteur of Cote d’Ivoire was alerted by possible cases of Yellow fever. Samples were sent to the French National Reference Center (FNRC) for Arboviruses that identified them as Dengue 3 genotype III (DENV-3 genotype III) isolates. In 2010, three travelers returning to Japan from Tanzania and Côte d’Ivoire developed a dengue fever belonging to the DENV-3 genotype III.31
The objective of the present report was to study the circulation of DENV-3 genotype III in Africa since 2008. This approach provides new insights on the spreading of DENV-3 genotype III viruses in Africa.
Virus isolation
Between 2008 and 2012, 15 DENV-3 strains with an African origin were isolated by the French National Reference Center for Arboviruses: 2 from Cote d’Ivoire in 2008, 2 from Cape Verde in 2008 and 2009, 1 from Togo in 2009, 1 from the Union of Comoros in 2010, 2 from Madagascar in 2010, 4 from Djibouti in 2011 - 2012 and 3 from La Réunion Island in 2012 (Table 1).
Single T25 C6/36 cells flask (2,6.106 cells) was inoculated with a mix made of 200 μl of serum with 2 ml of L-15 cell culture medium (Gibco), 2% FCS and 20 μl of heparin and filtered on a 22μm sterile filter. Culture flask was incubated at 28 °C. After 7 days, 2 ml of media was removed and added to a new T25 C6/36 cells flask. Culture flask was incubated at 28 °C, under 5% CO2 during 5 days. The supernatant was then removed and used for RT-PCR.
Reverse-Transcription and sequencing
Viral RNA was extracted from 140 μl of cell culture supernatant using QIAamp Viral RNA Mini Kits (Qiagen, Germany). RT-PCR and amplification was processed as previously described by Lanciotti et al.17 To generate two overlapping fragments covering the PrM/M and E genes region, primers pairs F1 (5’-GCTCCCCATGTCGGCATGGGACTGG-3')/ R1 (5’-CATCCCTTTGAGTTTCAATTTGTCCAT-3') (1541bp) and F2 (5’-CTAGGATCTCAAGAAGGAGCAATGCA-3’)/R2 (5’-CGCGGATCCATGGCTGTTGCCACTCTTTTGGGGGA-3') (865bp) were used in 2 separate reactions. These 2 overlapping fragments amplify a global region of 2272bp. RNA was first transcribed to cDNA using the Super Script III one-step RT-PCR kit (Invitrogen). The reaction mix was 25 µl of 2X reaction mix, 5 µl of template RNA, 1µl of sense primer 10µM (F1 or F2), 1µl of anti-sense primer 10 µM (R1 or R2), 2 µL of SuperScript III RT/Platinum Taq Mix and up to 50 µl of autoclaved distilled water according to manufacturer instruction. The cycle used was 56 °C during 30 min then 94 °C during 2 min followed by 40 cycles of 94 °C (15 s), 56 °C (30 s) and 68 °C (1 min 30). To finish, a final step of elongation at 68 °C during 5 min was performed.
PCR products were sequenced by Beckman’s Coulter Genomics, UK. The 15 E gene sequences are deposited in Genbank database under accession numbers listed as follows: KT187281, KT187282, KT187283, KT187284, KT187285, KT187286, KT187287, KT187288, KT187289, KT187290, KT187291, KT187292, KT187293, KT187294, KT187295 (Table 1).
Sequence dataset
Different datasets were constructed to perform phylogenetic analyses. For the determination of the DENV-3 genotype of the 15 strains, panels of 101 DENV-3 GenBank E gene sequences of 1479 bp of all genotypes were selected according to their geographic origin and date of isolation (Table 1).
Country |
Year |
Genotype |
Genbank Accession Number |
French Polynesia |
1994 |
I |
AY744685 |
Guangzhou |
2010 |
I |
JN009098 |
Indonesia |
1982 |
I |
DQ401690 |
Indonesia |
2010 |
I |
JF968101 |
Philippines |
2010 |
I |
JF968099 |
Taiwan |
1995 |
I |
DQ675519 |
Bangladesh |
2008 |
II |
JF968064 |
Cambodia |
2006 |
II |
EU448440 |
Myanmar |
2009 |
II |
JF968088 |
Thailand |
1973 |
II |
GQ868593 |
Thailand |
2000 |
II |
AB111082 |
Thailand |
2010 |
II |
JF968094 |
Benin |
2010 |
III |
AB690858 |
Bhutan |
2006 |
III |
FJ606692 |
Bhutan |
2006 |
III |
FJ606701 |
Bhutan |
2006 |
III |
FJ606702 |
Bhutan |
2007 |
III |
FJ606710 |
Bhutan |
2007 |
III |
FJ606711 |
Bhutan |
2007 |
III |
FJ606712 |
Brasil |
2002 |
III |
AY679147 |
Brasil |
2006 |
III |
EU617027 |
Brasil |
2006 |
III |
EU617029 |
Brasil |
2006 |
III |
EU617034 |
Brazil |
2006 |
III |
EU617035 |
Brazil |
2006 |
III |
EU617037 |
Brazil |
2006 |
III |
EU617038 |
Cambodia |
2000 |
III |
AB111081 |
Cape Verde |
2008 |
III |
KT187284 |
Cape Verde |
2009 |
III |
KT187285 |
Colombia |
2003 |
III |
GQ868572 |
Colombia |
2005 |
III |
FJ389916 |
Colombia |
2006 |
III |
FJ389915 |
Colombia |
2007 |
III |
FJ389914 |
Cote d'Ivoire |
2008 |
III |
AB447989 |
Cote d'Ivoire |
2008 |
III |
KT187283 |
Cote d'Ivoire |
2008 |
III |
KT187282 |
Djibouti |
2011 |
III |
KT187292 |
Djibouti |
2011 |
III |
KT187293 |
Djibouti |
2012 |
III |
KT187294 |
Djibouti |
2012 |
III |
KT187295 |
Guangzhou |
2009 |
III |
GU363549 |
Guangzhou |
2009 |
III |
HM466964 |
Guangzhou |
2009 |
III |
HM466966 |
Guangzhou |
2009 |
III |
JN662391 |
Guangzhou |
2010 |
III |
JN009093 |
Guangzhou |
2010 |
III |
JN009097 |
India |
2003 |
III |
AY770511 |
India |
2005 |
III |
JQ686077 |
India |
2005 |
III |
JQ922556 |
India |
2005 |
III |
JQ922557 |
India |
2007 |
III |
FJ644564 |
India |
2008 |
III |
GQ466079 |
India |
2009 |
III |
JQ686081 |
India |
2010 |
III |
JQ686079 |
India |
2011 |
III |
KP176703 |
India |
2011 |
III |
KP176704 |
Madagascar |
2010 |
III |
KT187290 |
Madagascar |
2010 |
III |
KT187291 |
Malaysia |
2010 |
III |
JF968107 |
Martinique |
1999 |
III |
AY099337 |
Mozambique |
1985 |
III |
FJ882575 |
Pakistan |
2006 |
III |
KF041238 |
Pakistan |
2006 |
III |
KF041249 |
Pakistan |
2006 |
III |
KF041256 |
Pakistan |
2006 |
III |
KF041257 |
Pakistan |
2006 |
III |
KF041259 |
Pakistan |
2007 |
III |
KF041255 |
Pakistan |
2008 |
III |
KF041254 |
Pakistan |
2009 |
III |
KF041258 |
Pakistan |
2013 |
III |
KM217132 |
Reunion |
2012 |
III |
KT187286 |
Reunion |
2012 |
III |
KT187287 |
Reunion |
2012 |
III |
KT187288 |
Samoa |
1986 |
III |
L11435 |
Saudi Arabia |
1997 |
III |
AM746228 |
Saudi Arabia |
2004 |
III |
AM746229 |
Saudi Arabia |
2004 |
III |
AM746231 |
Saudi Arabia |
2004 |
III |
AM746232 |
Saudi Arabia |
2014 |
III |
KJ830751 |
Singapore |
2005 |
III |
EU081198 |
Singapore |
2007 |
III |
JN030193 |
Singapore |
2012 |
III |
KX380841 |
Singapore |
2013 |
III |
KX224279 |
Singapore |
2013 |
III |
KX380842 |
Somalia |
2011 |
III |
KC848585 |
Somalia |
2011 |
III |
KC848586 |
Somalia |
2011 |
III |
KC848587 |
Somalia |
2011 |
III |
KC848588 |
Somalia |
2011 |
III |
KC848589 |
Sri Lanka |
1981 |
III |
L11431 |
Sri Lanka |
1985 |
III |
L11436 |
Sri Lanka |
1997 |
III |
GQ252674 |
Sri Lanka |
1989 |
III |
FJ882572 |
Sri Lanka |
1993 |
III |
FJ882573 |
Tanzania |
2010 |
III |
AB549332 |
Thailand |
2010 |
III |
JF968092 |
Thailand |
2010 |
III |
JF968098 |
Thailand |
2010 |
III |
JF968106 |
Togo |
2009 |
III |
KT187281 |
Togo/Benin/Burkina Faso |
2013 |
III |
KJ922394 |
Union of Comoros |
2010 |
III |
KT187289 |
Venezuela |
2000 |
III |
AY146766 |
Venezuela |
2007 |
III |
GQ868587 |
Yemen |
2010 |
III |
HQ336219 |
Zhejiang |
2009 |
III |
JF504679 |
Zhejiang |
2009 |
III |
GU721065 |
Zhejiang |
2009 |
III |
GU721066 |
Zhejiang |
2009 |
III |
GU721067 |
Zhejiang |
2009 |
III |
GU721069 |
Zhongshan |
2013 |
III |
KJ545473 |
Zhongshan |
2013 |
III |
KJ545474 |
Puerto Rico |
1963 |
IV |
AY146762 |
Brazil |
2010 |
V |
EF629370 |
China |
1980 |
V |
AF317645 |
Guangzhou |
2009 |
V |
HM466962 |
Philippines |
1956 |
V |
AB609590 |
Table 1 DENV-3 strains used in the study. New isolates are noticed in bold type
To compare the genetic relatedness of the partial virus genome, phylogenetic analyses were performed with a panel of 84 published DENV-3 genotype III sequences from 1986 up to 2014, and a DENV-3 genotype IV isolated in Puerto Rico in 1963 (AY146762) was used as an out group to root the tree. Before phylogenetic inference, datasets and multiple sequence alignments were thoroughly examined to eliminate misalignments and ensure correct framing of coding sequences leading to a 1479 bp sequence. Sequence alignments were performed by the Mega 5 program using the multiple alignment algorithms Clustal X.32
Phylogenetic analyses
The percentage of sequence identity was calculated using Bioedit v3.6 program. Sequencher® version 5.3 sequence analysis software (Gene Codes Corporation, Ann Arbor, MI USA) was used to analyze nucleotidic/amino-acid sequences. Maximum like hood phylogenetic tree was generated using the method implemented in the PAUP package.33 Model Test34 was used to determine the best model of substitution. The best fit to DENV-3 was the General Time Reversible + I + Γ model of substitution. Parcimony phylogenetic tree was also estimated using PAUP package.33
Phylogeny of the 15 strains isolated in Africa amongst DENV-3 strains
A total of 15 African strains from 7 different countries were isolated between 2008 and 2012 from African patients or from imported cases. Their E gene sequences were compared with 101 sequences representative of all five DENV-3 genotypes available in open databases. The phylogenetic analysis highlighted that the 15 African strains clustered within genotype III (Figure 1).

Figure 1 Phylogenetic tree of 116 DENV-3 strains based on E nucleotidic sequence (1-1479) using the Maximum Likelihood method based on the General Time Reversible + I + Γ model. Strains isolated in our laboratory, identified by *, belong to the genotype III. Node values were determined for 1,000 replicates. Branch lengths are correlated to the number of substitutions per site. Isolates are labeled as follows: country, date of isolation, genbank accession number.
DENV-3 includes five genotypes numbered from I to V that are mainly described in Asia and the Americas. Little is known about DENV-3 genotypes in Africa that could be explained by a reduced circulation in Africa or a lack of surveillance. In Africa, only genotype III has been reported. This genotype is highly likely to have been introduced in Mozambique in 1985 from India.17 The objective of the present study was to analyze DENV-3 genotype III in Africa in order to improve our understanding of its diversity. This study was realized on a limited number of samples as it mainly came from European travelers back from limited area in Africa. Effectively, returning travelers currently serve as sentinels for local outbreaks of dengue fever in African countries where surveillance system are inadequate.
Phylogeny of the 15 strains isolated in Africa amongst DENV-3 genotype III strains
We mainly focused on genotype III strains isolated in Africa using a parcimony analysis (Figure 2A). Phylogenetic analyses separated strains isolated in South America from strains isolated in Africa and Asia (Figure 2A). With the African-Asian group, all strains isolated in Africa from 2008 to 2012 are clustered within 5 subgroups, except strains from La Reunion isolated in 2012 that clustered within an Asiatic subgroup including a strain from Thailand in 2010.
Within the African-Asian group, Malagasy and Comorian strains grouped together with Tanzanian and Guanghzou strains (Figure 2B). Djiboutian strains formed a regional East- African phylogenetic group so as Somalian strains. The strain isolated in Yemen is phylogenetically different from the other African strains and is considered by itself as a separate subgroup. In West Africa, we identified a regional subgroup composed of strains isolated in Benin, Cape Verde, Ivory Coast and Togo, but also in China, in Guanghzou and Zhejiang in 2009 (Figure 2B).

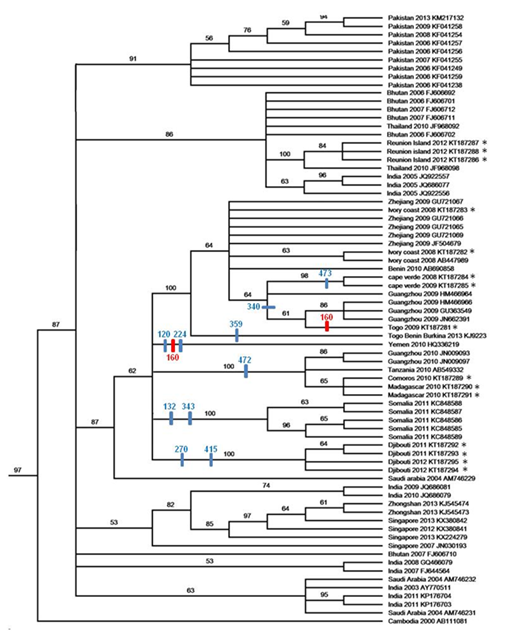
Figure 2 2A: Phylogenetic tree of 116 DENV-3 strains 1986- 2014 based on nucleotidic sequence of E (1-1479) using Parcimony analysis (Boostrap consensus tree). DENV-3 genotype IV isolated in Puerto Rico in 1963 (AY146762) was used as an out-group to root the tree. Node values were determined for 1,000 replicates. Branch lengths measured the number of substitutions per site. The 15 strains isolated in Africa between 2008 and 2012 are represented by*. 2B: Magnification of the part of the parcimony analysis. New sequences are noticed by *. On the branches of the tree, in blue, are noticed amino-acid positions of acquired non-synonymous mutations compared to the Saudi Arabia 2004 strain (G340E, M473L, K359R, Q120H, T224A, V489A, S472P, N270T, D415E, Y132H, K343R), and in red the mutation A160V which appeared independently in 2 lineages. Isolates are labeled as follows: country, date of isolation, genbank accession number.
The E gene is frequently used for phylogenetic analysis as it corroborates the phylogenetic analysis based on the sequences of the entire coding region.35 Phylogenetic analysis DENV-3 clearly separated strains isolated in South America from strains isolated in Africa and Asia that clustered together. This firm phylogenetic linkage between DENV-3 Genotype III strains collected in the same regional areas of Africa and Asia, was already described by Zaki et al.36
This study is the first report of DENV-3 presence in La Reunion as DENV-1 was the only serotype detected on the island since the dengue serotype surveillance started in 1987.37 The phylogenetic analyses (Figure 2B) revealed that strains from La Reunion isolated are probably not the result of a regional virus transmission (i.e. Madagascar or the Union of Comoros) as it shared 99.9% nucleotidic identity with a strain isolated in Thailand in 2010, as previously suggested by Larrieu et al.38
Tanzanian strains were phylogenetically linked to the strains isolated in the Union of Comoros, Madagascar and China (Guanghzou 2010), hence suggesting viral flux between the southeastern African coast and the islands across the Mozambique Channel. We can also assume that transmission from the Union of Comoros to Madagascar is a credible scenario due to the DENV-3 2010 Comorian epidemic.39,40 Intense human flows, animal movements and trade have been present for centuries between the triptych of Madagascar, the Comoros archipelago and the countries of the East African coast across the Mozambique Channel. This is not the first report of a new pathogen introduction in the Indian Ocean Islands from the African continent. Commercial trade has already affected the health status of the Union of Comoros in regards to many diseases, such as blackleg in 1970 and 1995, contagious ecthyma in 1999, East Coast fever in 2003 and 200441,42 and Rift Valley fever in 2011.43 The Tanzanian strain also shares a common ancestor with the strain isolated in China (Guangzhou) that have probably circulated during business travels.44 Regionally, current circulation of the virus through a large part of eastern Africa was evidenced by the identification of new DENV-3 strains in other African countries. Effectively, DENV-3 was identified in Eritrea in 2008 (Vapalahti, personal communication) and in Sudan in 201045 but no sequence is currently available. In West African countries (Togo/Benin/Burkina, Cote d’Ivoire, Togo and Cape Verde), we can observe a regional circulation of the virus, as suggested by the isolation in 2010 of a DENV-3 genotype III strain in Gabon which is phylogenetically linked to the strains from Cote d’Ivoire.46 Local/regional circulation of goods and population in this area could favor the importation of new strains and the diffusion of the virus. Identification of closely related strains isolated in China states the idea of a possible circulation with Western African countries probably by commercial exchanges. Effectively, the relationship between the increased economic investment and numbers of exported laborers to Africa from China and the increased number of malaria cases imported is well established.47,48 This strain is also phylogenetically linked to the ones responsible for the 2 epidemics in 2009 and in 2010, respectively in the Chinese provinces of Zhejiang and Guangzhou49 where only genotypes I and V were previously documented.44 Finally, diversity observed within the strains isolated in Africa over a 5-year time period evidence extensive virus exchanges associated with multiple virus introductions. These introductions are probably the consequence of trade but also of tourism.
Amino acid mutations associated with the circulation of the 15 strains
Figure 2B also presented amino acid mutations that occurred during circulation of the strains and evidenced an independent apparition of the same amino-acid mutation A160V in Yemeni and Togolese strains. Interestingly, this mutation appears many times independently, worldwide, in different DENV-3 genotypes. For example, strains from Cambodia in 2006 (EU448440), Bangladesh in 2008 (JF968064) and Thailand in 2010 (JF968094) belonged to the genotype II and also presented a valine instead of an alanine in position 160 of the E protein. Mutations on the E protein may have implications in the attachment to the cell membrane and the entrance of the virus in the cells.50,51 It could be associated with the virus’s flexibility towards its vector or host(s). Further investigations should be done to determine a possible mutation’s implication.
Evolution through adaptive genetic changes and introduction of newer viral strains by cross-border transmission play a major role in making dengue virus a successful pathogen of the tropics. It is fairly well established that such acquired changes affect the dengue disease severity.
With the addition of the first report on dengue virus type 3 genotype III in Africa in 1984, our results evidenced the circulation of the genotype over Africa since its first descriptions in Mozambique and Somalia between 1984 and 199318,19,30 and in Cameroon in 2006.52 Diversity of the strains analyzed suggests extensive virus exchanges associated with multiple virus introductions that are probably the consequence of trade and tourism. In 2014, a new strain originated from Thailand or Malaysia was detected in Saudi Arabia, pointing out the permanent risk of importation of new strains that may rapidly spread out over Africa and surrounding territories. A keen monitirong of DENV-3 genotype III circulation would be a major requirement for successful dengue surveillance and would enlighten our perception of the progression of dengue in Africa.
The authors are grateful to Toscane Fourié for her reviewing and advice about the English version of the manuscript.
None.

©2016 Monteil, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.