Journal of
eISSN: 2373-6453


Research Article Volume 4 Issue 1
Department of Microbiology and Immunology, The University of Western Ontario, Canada
Correspondence: C Yong Kang, Department of Microbiology and Immunology, Schulich School of Medicine and Dentistry, Siebens-Drake Research Institute, The University of Western Ontario, London, Ontario, N6G 2V4, Canada, Tel 1-519-661-3226, Fax 1-519-661-3359
Received: November 08, 2016 | Published: November 23, 2016
Citation: Kim GN, Wu K, An HY, Banasikowska E, Harding M, et al. (2016) Matrix Protein Gene Variants of Two Distinct Serotypes of rVSV Make Effective Viral Vectors for Prime-Boost Vaccination. J Hum Virol Retrovirol 4(1): 00125. DOI: 10.15406/jhvrv.2016.04.00125
Recently, we developed attenuated VSV vectors by introducing temperature sensitive (ts) mutations in the M gene in both VSV Indiana and New Jersey serotypes. The newly generated M gene mutants of rVSV vectors are rVSVInd(GML) with mutations of G21E, M51R, and L111F, rVSVNJ(GMM) with mutations of G22E, M48R, and M51R, and rVSVNJ(GMML) with mutations of G22E, M48R, M51R, and L110F. Our purpose was to examine the immunogenicity of the new ts M gene mutant of rVSVInd and attenuated M gene mutants of rVSVNJ as vaccine vectors against HIV-1 proteins. We generated attenuated rVSVs carrying HIV-1 gag, pol, and env genes. We immunized mice with various prime-boost vaccination regimens. CD8+ T cell responses and humoral immune responses in the vaccinated mice were examined. Priming with rVSVInd(GML)-gag, pol, or env gene of HIV-1 and boosting with rVSVNJ(GMM)-gag, pol, or env gene or rVSVNJ(GMML)-gag, pol, or env gene induced the strongest CD8+ cytotoxic T cell responses against the HIV-1 Gag, RT, and Env proteins. The same vaccination regimen also induced strong humoral immune responses against the HIV-1 Gag and Env proteins. We conclude that rVSVInd(GML) priming followed by rVSVNJ(GMM) boosting is the best vaccination regimen for optimum B cell and T cell adaptive immune responses against inserted foreign gene products when the newly attenuated rVSVInd and rVSVNJ are used.
Keywords:Vesicular stomatitis virus, Temperature sensitive mutants, HIV-1, Vaccine, Adaptive Immune Responses
VSV, Vessicular Stomatitis Virus; HIV, Human Immunodeficiency Virus; Ind, Indiana; NJ, New Jersey; HR, Heat Resistant; ELISA, Enzyme-Linked Immunosorbent Assay
In addition to three conventional vaccine types, such as live attenuated virus vaccine, killed whole-virus vaccine, and subunit vaccines, scientists have developed vaccines using recombinant viral vectors. Many different types of viruses are employed to deliver genes of interest. These viral vectors include poxviruses,1 adenoviruses,2,3 herpes viruses,4 alphaviruses,5 retroviruses6 and rhabdoviruses.7-9 The ideal recombinant viral vector system should be able to carry large size exogenous gene(s) with high levels of the inserted gene expression. In addition, it should have a broad range of hosts and must be safe.
A boost immunization is often necessary in order to induce the highest adaptive immune responses. For a prime-boost immunization regimen, the priming recombinant viral vector should be antigenically distinct from the boost vaccine vector. The priming vaccine vector will most likely induce antibodies which will neutralize the boosting vaccine vector, should one use the same vector for prime-boost vaccination. Therefore, ideally one should use two different recombinant viral vectors, which are antigenically distinct.
The vesicular stomatitis virus (VSV) offers an ideal delivery system for prime-boost vaccines. VSV is the prototype rhabdovirus, which replicates rapidly and has a wide host range. Both humoral and cellular immune responses against VSV are elicited in the animal host, like any other viral vector.10-12 VSV is neutralized by serotype specific antibodies against the viral surface glycoprotein G. VSV has two major serotypes; Indiana serotype (VSVInd) and New Jersey serotype (VSVNJ). VSVInd and VSVNJ show 50% amino acid homology in their glycoprotein G.13 However, antibodies raised against VSVInd do not neutralize VSVNJ.14 Therefore, other investigators have used VSVInd as a vaccine vector in which the glycoprotein gene of VSVInd was replaced with that of VSVNJ to minimize the problems arising from this immune response against the same viral vectors.8,15
Although VSVInd carrying the G protein gene of the VSVNJ serotype, or Chandipura as serologically distinct VSV, is useful in evading humoral immune response, it may not prevent cellular immune response which can be triggered by four other structural proteins: N, P, M, and L of VSV. Cellular immune responses against VSV proteins, other than the G protein, may result in incomplete immune responses against the antigen of interest. Our rVSVInd was generated from a cDNA clone of the HR strain of VSVInd16 and rVSVNJ was generated from a cDNA clone of the Hazelhurst strain of VSVNJ.17 The two serotypes of VSV demonstrate different capacities for inducing type I interferons (IFN-α/β). Generally, VSVNJ is a better inducer for type I interferons.18 The type I interferons act as stimulatory factors to induce both cellular and humoral immune responses.19 The generation of additional recombinant VSV from another serotype such as VSVNJ may increase the efficacy of using VSV as a live viral vaccine vector. Using both recombinant VSVInd and VSVNJ creates an effective viral vector system for the expression of foreign genes, which can be used to minimize problems associated with pre-existing immune responses against VSV itself.
Recently, we reported the attenuation of rVSVInd by introducing temperature-sensitive M gene mutations (G22E/L111F) and combining them with non-cytopathic M gene mutation (M51R). The newly generated rVSVInd is rVSVInd(G21E/M51R/L111F-GML), which is temperature-sensitive, assembly-defective rVSVInd. Similar mutations were introduced into the M gene of the New Jersey serotype of rVSV, and the resulting viruses were not temperature sensitive but were more attenuated from the non-cytopathic M gene mutant, rVSVNJ(M48R/M51R). The newly generated rVSVNJ are rVSVNJ(G22E/M48R/M51R-GMM) and rVSVNJ(G22E/M48R/M51R/L110F-GMML).16
We examined the efficacy of these newly attenuated rVSVs as vaccine vectors by inserting three different HIV-1 genes, gag, pol, and env. Here we report the induction of both humoral and cellular immune responses against HIV structural proteins, Gag, Pol, and Env using the dual serotype rVSV vaccine vectors. Recombinant VSVInd and VSVNJ carrying the gag, pol or env genes of HIV-1NL4-3 were constructed. Mice primed with rVSVInd carrying HIV structural protein genes followed by boost immunization with rVSVNJ carrying the same set of HIV-1 structural protein genes showed robust adaptive immune responses.
Cells
Baby hamster kidney cells (BHK21, ATCC®, CCL-10TM) were grown in DMEM (Invitrogen) containing 5% fetal bovine serum (FBS). African green monkey kidney cells Vero E6 (ATCC®, CRL-1586TM) were maintained in MEM (Invitrogen) containing 10% FBS and 1 mM sodium pyruvate. BSR T7/5 cell20 was obtained from Dr. K.K. Conzelmann. The BHK21 cells constitutively expressing the bacteriophage T7 RNA polymerase (BSR T7/5) were grown in DMEM containing 5% FBS and 500 µg/ml of G418 (Invitrogen). Splenocytes from vaccinated mice were isolated and kept in RPMI media with 10% FBS.
Construction of plasmids
The cDNA of the HIV-1 gag gene was synthesized by PCR using pUC19-gagWR27-TCE-EN21 as a template and primers Gag(F):5'-GGACGCGTGATGAACGATATGAAAAAAACTAACAGAATTC
AAAATGGGTGCGAGAGCGTCAATATTAA-3' and Gag(R): 5'-CGGCGCGTTACTTCCACACAGGTAC
CCCATAA-3'. The PCR products were inserted into the Mlu I site of prVSVInd(WT), prVSVInd(GML), prVSVNJ(WT), prVSVNJ(GMM), and prVSVNJ(GMML) at the G gene and L gene junction. The plasmids for the rVSV with the HIV-1 gag gene are prVSVInd(WT)-gag, prVSVInd(GML)-gag, prVSVNJ(WT)-HIV-gag, prVSVNJ(GMM)-gag, and prVSVNJ(GMML)-gag.
The cDNA of the HIV-1 env gene with a honeybee melittin signal peptide gene sequence was synthesized from a template, pNL4-3 (Dr. M. Martin, Division of AIDS, NIAID, NIH) by PCR using primers Env-mss(1F): 5´-CGGGCGGCCGCATGAAATTCTTAGTCAACGTTGCCCTTGTTTTTA
TGGTCGTATACATTTCTTACATCT ATGCGGCCACAGAAAAATTGTGGGTCACAGTCTATT-3´ and Env-mss(1R): 5´-CGGGCGGCCGCTTATAGCAAAATCCTTTCCAAGCCCTG-3´. The PCR product was cloned into pBluescript II-KS vector (Agilent Technologies) to generate the pBKS-Env-mss. VSV intergenic junction sequences were added to the front of HIV-1 Env-mss by PCR using primers Env-mss(2F): 5´-CGAGCTCACGCGTGATGAACGATATGAAAAAAACTAACA
GAATT CAAAATGAAATTCTTAGTCAACGTTGCCCTT-3´ and Env-mss (2R): 5´-CGAGCTCACGCGT
TTATA GCAAAATCCTTTCCAAGCCCTG-3´ and a template pBKS-Env-mss. The HIV-1 Env-mss gene with VSV intergenic junction sequences was cloned into pBKS to generate a pBKS-Env-mss-VSVIG. The HIV-1 Env-mss gene was cut with a restriction enzyme Mlu I and cloned into prVSVInd(GML) and prVSVNJ(GMM) to generate prVSVInd(GML)-HIV-env and prVSVNJ(GMM)-env. The cDNA of the HIV-1 pol gene with a VSV intergenic junction sequence was synthesized by PCR using primers Pol(F): 5´-CGAGCTCACGCGTGATGAACGATATGAAAAAAACTAACAGAATTCAAAATGTTTTTTTAGGGAA
GATCTGGCCTT and Pol(R): 5´-GAGCTCACGCGTTTAATCCTCATCCTGTCTACTTG-3´ and pNL4-3 as a template. The HIV-1 pol gene with a VSV intergenic junction sequence was cloned into a pBluescript II KS vector to generate pBKS-HIV-1 pol. The pol gene was cut with a restriction enzyme Mlu I and cloned into the prVSVInd(GML) and prVSVNJ(GMM) to generate prVSVInd(GML)-HIV-pol and prVSVNJ(GMM)-pol.
Recovery of recombinant VSV by reverse genetics
The rVSVs were recovered as described previously17 by VSV reverse genetics from the plasmids of prVSVInd(WT)-gag, prVSVInd(GML)-gag, prVSVNJ(WT)-gag, prVSVNJ(GMM)-gag, prVSVNJ(GMML)-gag, prVSVInd(GML)-env, prVSVNJ(GMM)-env, prVSVInd(GML)-pol, and prVSVNJ(GMM)-pol.
Viral preparation and titration
The newly generated rVSVs were purified three consecutive times by plaque picking, and were amplified for a larger volume of stock viruses. The plaque picked viruses were first amplified in a small-scale 2 ml culture of BHK21 cells and the viruses were titrated using a monolayer culture of Vero E6 cells by plaque assay. The cells infected with the mutant viruses for the plaque assay were incubated at 31 °C. For a larger viral stock, BHK21 cells were infected with an MOI of 0.1, incubated at 31 °C, and the culture media was harvested at 20 hours post-infection.
Determination of protein expression
In order to examine the expression of VSV proteins and inserted HIV-1 gene products, Gag, gp160, and reverse transcriptase (RT), proteins from 10 µg cell lysate of infected cells were separated by SDS-PAGE, and then transferred to the PVDF membrane (Immobilion, Millipore). The VSV proteins and HIV-1 proteins were detected by Western blot analysis using ECL Plus Western blotting Detection reagents (Amersham Biosciences). VSV proteins were detected by using rabbit serum against the total protein of VSV.22 The HIV-1 Gag protein was detected by using the HIV-1 p24 monoclonal antibody (183-H12-5C, NIH Cat#3537). The HIV-1 Env protein was detected by using the goat anti-HIV-1 gp120 polyclonal antibody (BIODESIGN). HIV-1 RT products were detected by using the mouse anti-HIV-1 RT monoclonal antibody (5B2B2, NIH Cat#11338).
Vaccination of mice with rVSV
Animal care and procedures were compliant with the Animal Care and Use Committee Guidelines of the University of Western Ontario. Six-week old female Balb/C mice (Charles River Laboratories) were lightly anesthesized with isoflurane (Baxter Corp, Mississauga, Ontario, Canada). The mice were injected intramuscularly in the hind leg with 50 µl of rVSVs of appropriate viral titre, which was diluted in serum-free DMEM. The vaccinated mice were housed in microisolator cages (three mice/cage) with a ventilated rack system. The vaccinated mice were weighed weekly until the mice were euthanized at the end of the fourth week of the study. For the prime-boost vaccination studies, mice were boost immunized three weeks after the prime immunization. The mice were euthanized one week after the boost immunization to collect their spleens and serum.
Peptide specific CD8+ T cell activation
In order to stimulate T cells in splenocytes, 100µl of 2X concentrated mixture of costimulant, anti-mouse CD28 (2 µg/ml, BD PharmingenTM) and an antigen specific peptide (20 µg/ml in dimethyl sulfoxide) were added to a 96-well cell culture plate. The HIV-1 Gag antigen specific peptide Gag: NH2-AMQMLKETI-COOH,23 gp160 specific peptide Env P18: NH2-RIQRGPGRAFVTIGK-COOH,24,25 RT specific peptides RT464: NH2-ILKEPVHGVYYDPSKDLIAE-COOH,26 and VSV nucleocapsid (N protein) antigen specific peptide N275: NH2-MPYLIDFGL-COOH27 were synthesized at GenScript (Piscataway, NJ, USA). Isolated splenocytes from the vaccinated mice (100 µl of 1X106 cells) were added to a 96-well plate that contained virus-specific peptides and anti-CD28. The splenocytes were incubated at 37 °C for 2 hrs and 10 µl of 1:50 diluted protein transport inhibitor Brefeldin A (BD GolgiPlug™, BD Bioscience) was added to the 200 µl of splenocytes in stimulation. The plate was incubated for an additional three hours, and the splenocytes were washed once in PBS/1% BSA. CD8 on the T cells were stained with anti-mouse CD8-FITC (BD PharmingenTM) at 4 °C for 30 minutes. The CD8 stained cells were washed twice with PBS/1% BSA and permeabilized with 75 µl of fixation/Permeabilization solution (BD Cytofix/CytopermTM plus) at 4 °C for 20 minutes. The permeabilized cells were washed twice with BD Perm/Wash™ Buffer and the cells were stained with anti-mouse IFN-g-APC (BD PharmingenTM). The stained cells were washed twice in Perm/WashTM buffer and were resuspended in 175 µl of PBS/1% BSA. The stained cells were identified using a FACS Calibur flow cytometer (BD Biosciences) and FlowJo software (Tree Star Inc., Ashland, OR). The data is expressed as an average % CD8+IFNγ+ (+/- standard deviation of the mean) for each vaccine.
ELISA
For the ELISA against HIV-1 Gag, a 96-well ELISA plate (R and D systems; Part# 992427) was coated with recombinant p55 Gag protein (Thermo Scientific; Cat# RP4921) at a concentration of 125 ng/well in PBS. For the ELISA against HIV-1 gp120, 96-well ELISA plate (R and D systems; Part# 992427) was coated with the HIV-1 gp140 trimer (NIH AIDS Reagent program; Cat# 12026) at a concentration of 250 ng/well in PBS. The mouse sera were diluted either at 1:100 or 1:400 with a blocking buffer (R and D systems; Part# 840149). The antibody bound to the antigen p55 Gag protein or gp140 was detected with a secondary antibody, sheep anti-mouse IgG-HRP (Amersham Bioscience; Cat# NA931V). The enzymatic activity of HRP was detected by adding substrates, a mixture of hydrogen peroxide (R and D systems; Part# 895000) and tetramethylbenzidine (R and D systems; Part# 895001). The OD of each sample was read at a wavelength of 450 nm with the microplate reader (Bio-Rad; model 550).
Sample group data were analyzed by a two-sided independent sample t test using the statistics software, Graph Pad Prism version 6. A p value of <0.05 was considered as statistically significant.
The attenuated rVSVs with the HIV-1 gag, pol, or env genes expressed proteins equally well at both 31 °C and 37 ᵒC
The M gene mutants of rVSVInd [rVSVInd(GML)] and rVSVNJ [rVSVNJ(GMM) and rVSVNJ(GMML)] showed increased safety of the rVSVs in mice.16 In addition to safety of rVSVs for human use, the potency of rVSV to induce strong immune responses against inserted gene products is important for a vaccine vector. In order to examine the efficacy of the new M gene mutants of rVSV to induce both humoral and cellular immune responses in vivo, HIV-1 structural protein genes, gag, env, and pol were inserted into the G gene and L gene junction in the full-length cDNA clones of wild type and the mutants of rVSV: rVSVInd(GML), rVSVNJ(GMM), and rVSVNJ(GMML) (Figure 1(a)). When expressed in vitro, unprocessed HIV-1 Gag proteins form virus like particles (VLP) and the VLP are secreted from the cells.28 Therefore, the HIV-1 Gag protein was a suitable protein to express from the new M gene mutants of rVSV to examine both cellular and humoral immune response. In addition, the full-length HIV-1 pol and env genes were inserted into both VSVInd [rVSVInd(WT) and rVSVInd(GML)] and VSVNJ [rVSVNJ(WT), rVSVNJ(GMM), and rVSVNJ(GMML)] in order to examine the induction of CD8+ T cell responses and humoral immune responses.
The expression of the Gag precursor protein P55, reverse transcriptase (RT) from pol gene, and gp160 from env gene were examined by Western blot analysis as described in Materials and Methods. In order to analyze the expression of HIV-1 proteins, BHK21 cells were infected with an MOI of six of the rVSVs and incubated at 31 °C and 37 °C, and cell lysates were prepared at six hours post-infection. rVSVInd(GML)-gag, rVSVNJ(GMM)-gag, and rVSVNJ(GMML)-gag expressed high levels of Gag protein at a permissive temperature of 31ᵒC (Figure 1(b)) and at a semi-permissive temperature of 37 ᵒC (Figure 1(c)), although Gag proteins from rVSVNJ(GMML) were comparatively lower than that from the rVSVInd(GML) and rVSVNJ(GMM). rVSVNJ(GMM)-env expressed gp160 slightly better than rVSVInd(GML)-env did (Figure 1(d)) at both 31 ᵒC and 37 ᵒC. The RT dimer, P66 and P51, were detected similarly in cells infected with rVSVInd(GML)-pol and rVSVNJ(GMM)-pol (Figure 1(e)). The expression levels of Gag, RT, and gp160 from these rVSVs were similar at both 31°C and 37 °C.
Figure 1 Expression of HIV-1 gene from the rVSVInd(GML), rVSVNJ(GMM), and rVSVNJ(GMML). (a) Cloning of HIV-1 gag, pol, and env genes into the rVSVInd and rVSVNJ. The HIV-1 gag, pol, and env genes were inserted into the junction of the G gene and L gene in the full-length cDNA clones of wild type and GML of rVSVInd and wild type, GMM, and GMML of rVSVNJ. (b&c) Expression of HIV-1 Gag from the rVSVs at 31 °C and 37 °C. (d) Expression of HIV-1 gp160 from the rVSVs at 31 °C and 37 °C. (e) Detection of HIV-1 RT products from the rVSVs at 31 °C and 37 °C. The expression of Gag protein from the rVSVs was examined by Western blot analysis using monoclonal antibody against HIV-1 P24.
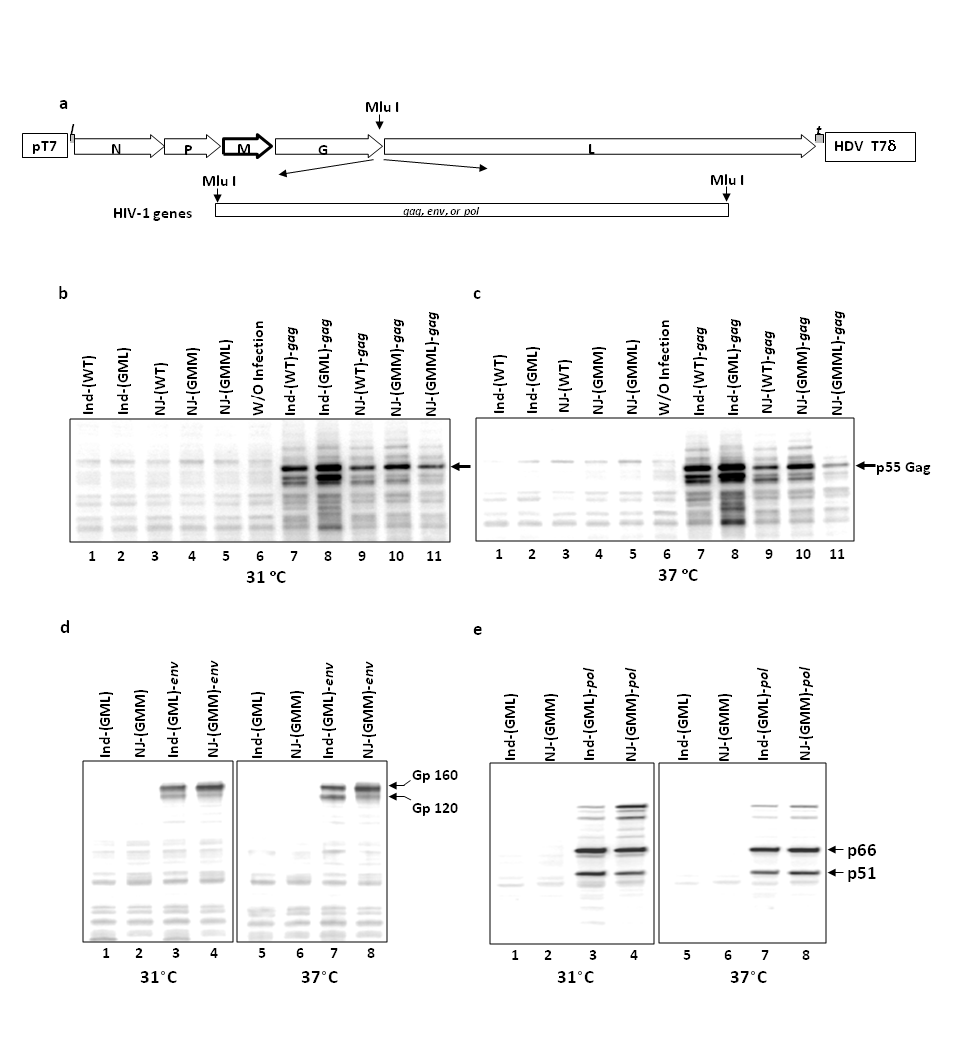
Figure 1 Expression of HIV-1 gene from the rVSVInd(GML), rVSVNJ(GMM), and rVSVNJ(GMML). (a) Cloning of HIV-1 gag, pol, and env genes into the rVSVInd and rVSVNJ. The HIV-1 gag, pol, and env genes were inserted into the junction of the G gene and L gene in the full-length cDNA clones of wild type and GML of rVSVInd and wild type, GMM, and GMML of rVSVNJ. (b&c) Expression of HIV-1 Gag from the rVSVs at 31 °C and 37 °C. (d) Expression of HIV-1 gp160 from the rVSVs at 31 °C and 37 °C. (e) Detection of HIV-1 RT products from the rVSVs at 31 °C and 37 °C. The expression of Gag protein from the rVSVs was examined by Western blot analysis using monoclonal antibody against HIV-1 P24.
The immunizations with the rVSVInd(GML)-gag priming followed by rVSVNJ(GMML)-gag boosting induce better immune responses than vice versa
It has been demonstrated that in the prime-boost vaccination regimen, avoiding neutralization of the boost virus by using the viral surface glycoprotein of another serotype is critical for the successful induction of strong immune responses.29,30 We examined the cell-mediated and humoral immune responses against genes of interest induced by two completely separate serotypes of attenuated rVSV in the prime-boost vaccination regimen. We compared the immune responses between regimens of the same serotype vs. two different serotypes for prime-boost vaccination as well as the immune responses between regimens using wild type rVSV vs. the mutant rVSVs.
We examined the immune responses against the HIV-1 Gag protein expressed from rVSVs by vaccinating six Balb/c mice per group as described in Materials and Methods and Figure 2. Mice were grouped according to vaccine vector types (wild type vs. mutant) and regimen, e.g., priming and boosting with the same serotype of rVSV, or by alternating the two serotypes for priming and boosting (Figure 2). The mice were prime-vaccinated intramuscularly with 5X106 PFU of rVSVs at the age of six weeks. Three weeks after priming, the mice were boost-vaccinated with the same dose of rVSVs. One week after boost vaccination, splenocytes and sera were collected for determination of the HIV-1 Gag-specific CD8+ T cell immune responses and anti-Gag antibody responses.
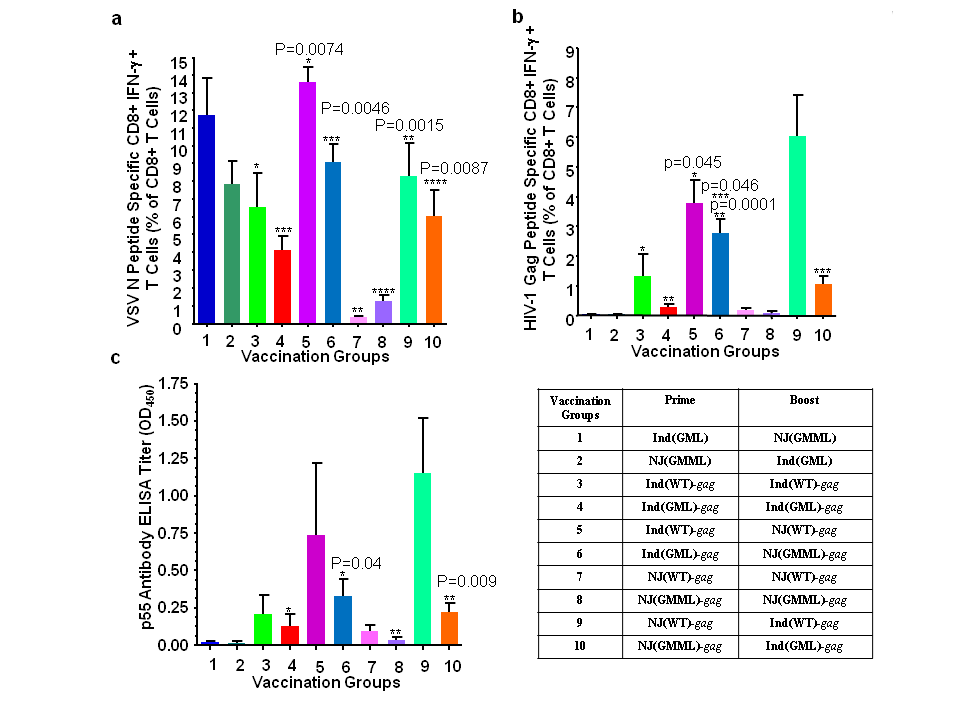
Figure 2 Determination of the best regimen for prime and boost immunization using rVSVInd and rVSVNJ. (a) VSV N protein specific activation of CD8+ T cells. (b) HIV-1 Gag protein specific activation of CD8+ T cells. Six mice/group were prime-vaccinated intramuscularly with 5X106 PFU of rVSVs at the age of six weeks. Three weeks after the priming, mice were boost-vaccinated with the same dose of rVSVs. A week after the boost vaccination, spleens and sera were collected for the HIV-1 Gag specific CD8+ T cell immune responses and humoral immune responses. (c) Antibody production against HIV-1 Gag. Generation of HIV-1 Gag specific antibody was examined with the serum collected at one week after boost immunization. The Gag specific antibody titre was determined by the indirect enzyme-linked immunosorbent assay (ELISA) with recombinant p55 Gag protein at a concentration of 125 ng/well. The mouse serum was diluted 1:100. The error bar represents standard deviation of the mean. The P values (*, **, ***, ****) were computed by using a two-sided independent sample t test.
CD8+ T cells stimulated by interacting with MHC I molecules loaded with peptides on the antigen presenting cells enhances the secretion of interferon-g (IFN-g).31 These peptide specific CD8+ T cells in splenocytes against VSV N proteins and HIV-1 Gag proteins were stimulated (Figure 2(a) and 2(b)). The splenic CD8+ T cells from groups 1 and 2, which were vaccinated with mutants of rVSVInd and rVSVNJ without the HIV-1 gag gene, were not stimulated by the HIV-1 Gag peptide, thereby demonstrating the specificity of CD8+ T cell stimulation with the HIV-1 Gag peptide. Prime and boost immunization by alternating two serotypes of wild type rVSV(WT) or two serotypes of rVSV mutants induced stronger CD8+ T cell immune responses against VSV N protein as well as HIV-1 Gag proteins, compared to a prime-boost vaccination with single serotype of rVSV as seen in groups 1, 2, 5, 6, 9, and 10 (Figure 2(a) and 2(b)). When vaccination regimens with the M mutants of rVSV were compared, priming with rVSVInd(GML)-gag and boosting with rVSVNJ(GMML)-gag induced anti-HIV-1 Gag CD8+ T cell responses better than the regimen with rVSVNJ(GMML)-gag priming followed by rVSVInd(GML)-gag boost (Figure 2(b), group 6 vs. group 10).
A humoral immune response against HIV-1 Gag was examined by ELISA using the serum collected a week after the boost immunization. The HIV-1 Gag protein specific antibody responses were induced the best when two serotypes of rVSVs, either wild type or mutant rVSVs with HIV-1 gag were alternated for prime and boost immunization as it was shown in HIV-1 Gag specific CD8+ T cell responses (Figure 2(c), groups 5, 6, 9, and 10). When vaccinated with the combined M gene mutants, mice showed slightly better humoral immune responses against the HIV-1 Gag protein after priming with the rVSVInd(GML)-gag and boosting with rVSVNJ(GMML)-gag than vice versa, but this difference was not statistically significant (Figure 2(c) group 6 vs. group 10). These results demonstrated that priming with rVSVInd(GML) and boosting with rVSVNJ(GMML) induced better CD8+ T cell immune responses compared to rVSVNJ(GMML) priming followed by rVSInd(GML) boosting, and similar levels of humoral responses were induced by alternating VSVInd(GML) and rVSVNJ(GMML), regardless of the order of serotypes.
Immunization with rVSVInd(GML) and rVSVNJ(GMM) vectors induces good adaptive immune responses against HIV-1 structural proteins
Testing various vaccination regimens with the new M gene mutants demonstrated that priming with rVSVInd(GML) and boosting with rVSVNJ(GMML) worked best to induce HIV-1 Gag specific CD8+ T cell responses and antibody responses (Figure 2). However, the immune responses were not as good as with wild type rVSVInd and rVSVNJ as vaccine vectors. Therefore, we examined whether or not we could enhance the cellular and humoral immune responses by increasing infectious doses of the vaccine vectors.
The rVSVNJ(GMML) and another M gene mutant of rVSVNJ, rVSVNJ(GMM) showed the same level of attenuation in vitro and in vivo,16 however the immunogenicity of rVSVNJ(GMM) against the inserted gene product had not been determined. Therefore, we included rVSVNJ(GMM) to examine and to compare the immunogenicity of these two rVSVNJ as boosting vectors. CD8+ T cell responses against HIV-1 Gag was better with the rVSVNJ(GMM) as a boosting vector compared to that with rVSVNJ(GMML) (Figure 3); therefore, we chose rVSVNJ(GMM) as the boosting vector for a prime-boost vaccination regimen with our M gene mutants of rVSV.
A viral infectious dose of 5X109 PFU induced the best cellular and humoral immune responses (Figure 3(b) and 3(c)). Immunization with rVSVNJ(GMM) as a boosting vector showed more dose dependant cellular immune responses. A dose of 5X108PFU showed clearly the enhanced cellular immune responses compared to the lower doses (Figure 3(b)). Therefore, for further experiments with rVSV expressing HIV-1 proteins, we immunized mice with 5X108PFU as a vaccine dose, which, we thought, would induce good immune responses.
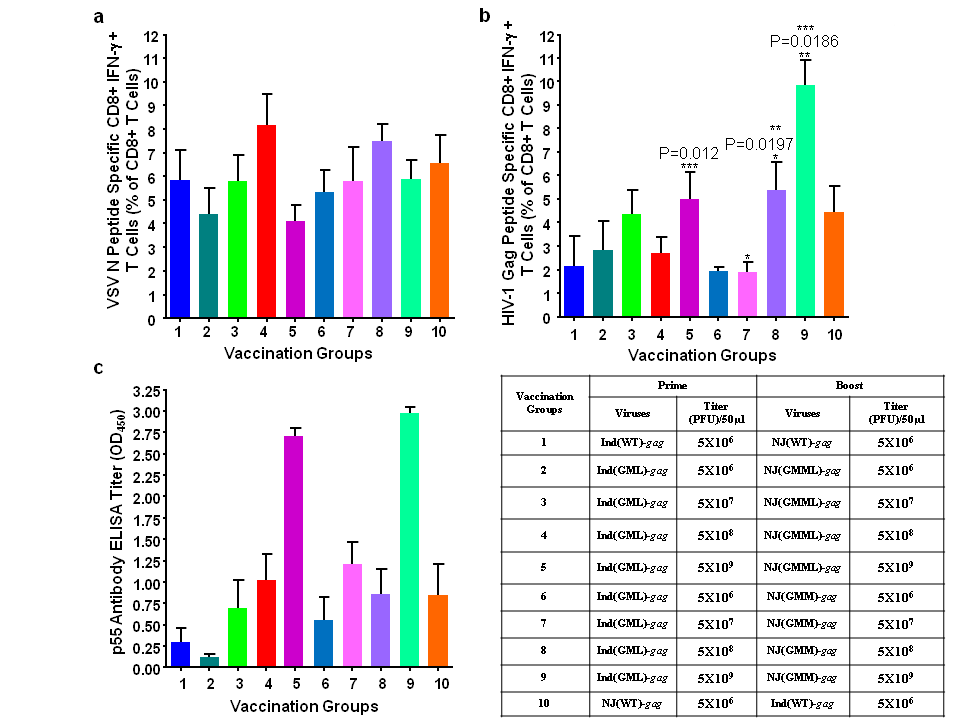
Figure 3 Immunization studies with increasing doses of rVSVInd(GML), rVSVNJ(GMM), and rVSVNJ(GMML) with HIV-1 gag. Six mice/group were prime and boost immunized with various doses ranging from 5X106 PFU/dose to 5X109 PFU/dose. The splenocytes and sera from the immunized mice were analysed as described in Figure 2. (a) VSV N protein-specific CD8+ T cell activation. (b) HIV-1 Gag protein-specific CD8+ T cell activation. (c) HIV-1 Gag protein specific antibody production measured by ELISA. The P values (*, **, ***, ****) were computed by using a two-sided independent sample t test.
With the optimized doses of immunization in mice, we examined immune responses against other HIV-1 proteins, such as gp160 and pol gene products, RT p51 and p66. Mice were prime-immunized with rVSVInd(GML) carrying HIV-1 genes and boost-immunized with rVSVNJ(GMM) with the same HIV-1 genes as shown in Figure 4. Mice were immunized with 5X108PFU of single virus or total of 1.5X109 PFU of three viruses (Figure 4). Splenocytes and sera were collected one week after the boost immunization. CD8+ T cell responses were peptide specific in all mice groups, as shown by the positive responses to peptides representing T cell epitopes of the HIV-1 proteins in groups 2, 3, 4, and 5 (Figure 4). Mice immunized with rVSVs expressing Gag, Env, and RT induced peptide specific CD8+ T cell immune responses in groups 2, 3, and 4 with different degrees of CD8+ T cell stimulation. CD8+ T cells against peptide Env P18 was highly efficient with about 19% CD8+ T cell stimulation in group 3, although we used a single Env-specific peptide (Figure 4(c)). About 5% of CD8+ T cells were stimulated against the Gag protein (Figure 4(b)) and 2% of CD8+ T cells were stimulated against the RT (Figure 4(d)). Compared to the single virus immunization in groups 2, 3, and 4, the mixed immunization with three viruses induced weaker CD8+ T cell immune responses against Gag and RT. The results of immunization against HIV-1 Gag, Env, and RT demonstrated that our rVSVInd and rVSVNJ prime-boost immunization strategy could induce specific CD8+ T cell immune responses against various viral proteins.
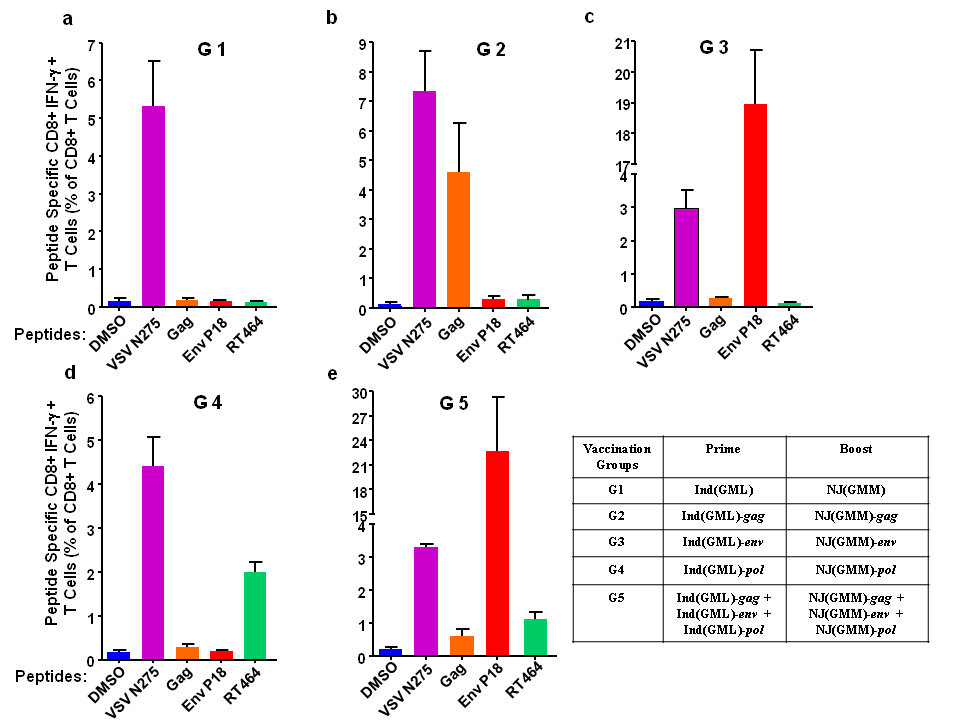
Figure 4 HIV-1 peptides specific CD8+ T cells induced by immunization with rVSVInd(GML) and rVSVNJ(GMM) expressing HIV-1 proteins. Six mice/group were primed with 5X108 PFU/dose of rVSVInd(GML) expressing HIV-1 Gag (b), Env (c), or RT (d) and boost immunized with 5X108 PFU/dose of rVSVNJ(GMM) expressing HIV-1 Gag, Env, or RT. The results were compared to those from mice immunized with rVSVInd(GML) and rVSVNJ(GMM) without HIV genes (a) and to those from mice immunized with all three rVSVs expressing HIV-1 proteins. The splenocytes from the immunized mice were analysed as described in Figure 2.
The humoral immune responses against HIV-1 Gag and Env proteins were examined by ELISA (Figure 5). The Gag precursor protein P55 was used as an antigen to detect antibodies against the Gag protein, and gp140 trimer was also used as an antigen to detect antibodies against the Env protein, as described in Materials and Methods. The production of the Gag antibody was significantly higher in groups 2 and 5 than that of group 1 (Figure 5(a)). Immunization with a single recombinant virus (group 2) rather than mixed viruses (group 5) induced better immune responses. Similar antibody titres against the Gag between groups 1, 3, and 4, which were not immunized with rVSVs expressing the Gag protein, indicated a slight cross reactivity between VSV proteins and HIV-1 Gag proteins. Env protein specific antibodies were generated only in groups 3 and 5, which were immunized with a rVSV expressing Env protein alone, or together with rVSVs expressing Gag and rVSV expressing RT (Figure 5(b)). The results demonstrated that the proteins expressed from the rVSVInd(GML) and rVSVNJ(GMM) and secreted from the infected cells could induce humoral immune responses in mice. Although it may not be statistically significant, it appeared that mice immunized with a single rVSV expressing Env protein (group 3) induced slightly better humoral immune responses than the mice immunized with mixed rVSVs (group 5). The results demonstrated that our new attenuated M gene mutants, rVSVInd(GML) and rVSVNJ(GMM) could induce Gag, RT, and Env protein specific CD8+ T cell responses and Gag and Env protein specific humoral immune responses.
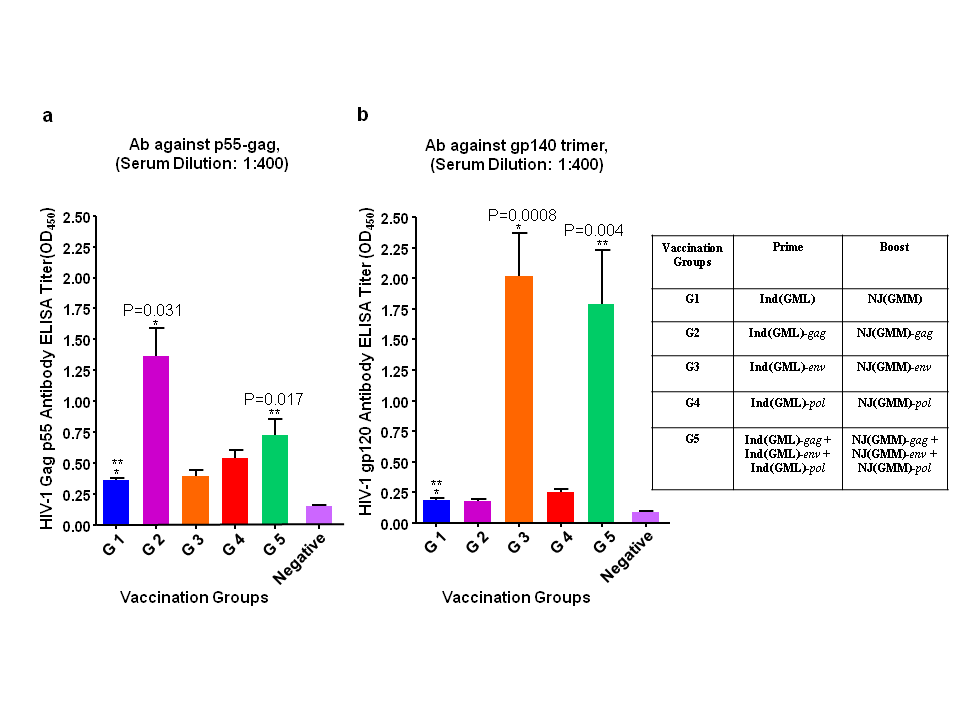
Figure 5 Humoral Immune responses induced by immunization with rVSVInd(GML) and rVSVNJ(GMM) expressing HIV-1 proteins. Mice were immunized as described in Figure 4. HIV-1 Gag p55 specific (a) and gp120 specific (b) antibody production was measured by ELISA. The P values (*, **, ***, ****) were computed by using a two-sided independent sample t test.
Our combined M gene mutants of rVSV are not deleted in any genes as compared to other known assembly-defective replication competent rVSV vectors. Our system uses rVSV with the full-length genome which assembly and release is reduced at a normal body temperature of 37 ᵒC. Therefore, our rVSV system does not require any complementary cell lines to provide the function of missing genes. Here, we tested the immunogenicity of these rVSVs with various immunization regimens against the genes of interest, HIV-1 gag, pol, and env genes expressed from the rVSVs.
It is generally considered that a higher dose of antigen increases the chances of naïve B and T cells to contact the antigen for activation.32 Higher doses of the live vaccine vector also affects CD8+ T cell immune responses by attracting a greater number of naïve CD8+ T cells to immune responses. This results in the activation of more antigen specific effector T cells and differentiated memory CD8+ T cells.33 The temperature sensitivity of rVSVInd(GML) for viral assembly should not affect the expression of VSV genes and genes of interest at a physiological temperature of 37 °C. Our rVSV vectors, the temperature-sensitive rVSVInd(GML) and rVSVNJ(GMM), and rVSVNJ(GMML) expressed similar levels of VSV proteins and HIV-1 Gag protein at both 31 °C and at 37 °C (Figure 1).
It has been demonstrated that prime-immunization of mice with the rVSVInd and boosting with the G gene mutant of other serotypes, such as New Jersey or Chandipura, induced better humoral immune responses.8 The results of our immunization studies with two antigenically distinct serotypes of VSV with the wild type M gene or mutant M gene of rVSVInd and rVSVNJ showed that prime and boost immunization with either rVSVInd priming and rVSVNJ boosting, or vice versa, worked better than the prime-boost with the same serotypes of rVSV. The order of the VSV serotypes for the prime-boost regimen to induce the best CD8+ T cell immune responses against the VSV N protein and HIV-1 Gag protein was not the same for both the wild type and M gene mutant rVSVs. HIV-1 Gag specific CD8+ T cell responses and humoral immune responses were better induced with the M gene mutants of rVSVInd priming and rVSVNJ boosting, while it was the opposite with the wild types of rVSVInd and rVSVNJ (Figures 2(b) &2(c)). From our experience with expressing foreign genes from the rVSVs, the expression level of the inserted genes and replication ability of the viruses in vitro vary depending on the inserted genes of interest. These characteristics of the rVSV expressing foreign genes and order of the serotypes of rVSV for the prime-boost regimen would affect various degrees of immune response. Using two serotypes of VSV is crucial to induce the best immune responses, but the order of the serotypes for prime-boost immunization may need to be determined empirically whenever new vaccines using the rVSV vaccine vector are generated.
The immunization dose of live vaccines may well influence the number of activated T cells and B cells against the antigen. In our study, we used rVSVInd(GML)-gag as a priming vaccine and matched it, either with the rVSVNJ(GMM)-gag or with the rVSVNJ(GMML)-gag, as a boosting vector to examine which M gene mutant of rVSVNJ vector stimulates better immune responses with the increasing doses. CD8+ T cell immune responses against the VSV N protein were not significantly different among all the groups with various doses of the rVSV vaccines, likely because of sufficient expression of VSV N proteins to activate a good number of naïve CD8+ T cells with the lowest dose of 5X106 PFU. Between the two M gene mutants of rVSVNJ, rVSVNJ(GMM)-gag and rVSVNJ(GMML)-gag as a boosting vector, rVSVNJ(GMM)-gag was more dose dependent than the rVSVNJ(GMML)-gag for the CD8+ T cell immune responses. The differences in the activation of the CD8+ T cells by the rVSVNJ(GMM)-gag and by the rVSVNJ(GMML)-gag could be a result of the differences in Gag protein expression by these two rVSVNJ.
The CD8+ T cell immune responses against the immunodominant Env p18 peptide were strongly induced in mice (Figure 4(c)&4(e)). Because the Env p18 peptide specific CD8+ T cell stimulation occurs in humans as well as in mice,24,34 it is anticipated that our immunization regimen using rVSVInd(GML)-env and rVSVNJ(GMM)-env could also induce strong CD8+ T cell immune responses in humans against the same region in the Env protein. Peptide specific CD8+ T cell responses against the Gag protein and RT were weaker when mice were immunized at the same site with mixed preparations of more than one rVSVInd(GML) or rVSVNJ(GMM) with HIV-1 genes than when mice were immunized with a single virus (Figure 4, group 2 and 4 vs. group 5). If multiple rVSVs are to be used for multiple antigens by a single immunization, we may need to optimize the number of doses and distribution of injection sites on the vaccinee.
Our new attenuated rVSVs; rVSVInd(GML), rVSVNJ(GMM), and rVSVNJ(GMML) demonstrated that they could be a safe vaccine vector with good expression of gene of interests. We will examine our rVSV with HIV-1 gag and env genes for the induction of human specific immune responses in humanized mice such as BLT mice.35 Although we may need to optimize viral doses and order of the two serotypes of rVSVs for human again, we believe that these rVSV vectors will be effective vaccine vectors for preparation of vaccines against various human viral and bacterial infections, such as infections by Ebola virus, hepatitis C virus, MERS coronavirus, mycobacterium tuberculosis. We will test our new VSV vectors for these human viral and bacterial vaccines as well.
The attenuated rVSV vaccine vectors expressed HIV-1 Gag, RT, and gp160 well and the level of exprssion was similar at both 31 ᵒC and 37 ᵒC. Prime and boost immunization by alternating two serotypes of rVSV induced stronger CD8+ T cell immune responses against VSV N protein as well as HIV-1 Gag proteins than using single serotype of rVSV. The results of immunization against HIV-1 Gag, Env, and RT demonstrated that our rVSVInd and rVSVNJ prime-boost immunization strategy could induce specific CD8+ T cell immune responses against various viral proteins. The results demonstrated that our new attenuated M gene mutants, rVSVInd(GML) and rVSVNJ(GMM) could induce Gag, RT, and Env protein specific CD8+ T cell responses and Gag and Env protein specific humoral immune responses.
This study was partially supported by an IRAP grant from the National Research Council of Canada and by a research contract from Sumagen Canada Inc. BSR T7/5 cells constitutively expressing bacteriophage T7 RNA polymerase was a kind gift of Dr. K.K. Conzelmann. We thank the AIDS Research and Reagent Program of the US National Institute of Health (NIH) for their support. We are grateful to Rosanne Kang and Doris Hall for editing the manuscript.
None.

©2016 Kim, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.