Journal of
eISSN: 2377-4312


Research Article Volume 3 Issue 2
Veterinary specialist institute Nis, Serbia
Correspondence: Marija Manic, Veterinary specialist institute Nis, Milke Protic bb, Nis, Serbi, Tel 381 64 0604 965
Received: February 19, 2016 | Published: March 8, 2016
Citation: Manic M, Nikolic S, Marjanovic V, et al. Resistance testing of Salmonella spp. bacteria isolated from samples derived from poultry on antimicrobial drugs. J Dairy Vet Anim Res. 2016;3(2):67-71. DOI: 10.15406/jdvar.2016.03.00074
In accordance with the Ordinance on establishing measures for the early detection, diagnosis, prevention of spreading, suppression and eradication of poultry infection by certain Salmonella serotypes, antimicrobial agents are also applied in controlling salmonellas is of poultry, and the very treatment may lead to development of resistance to different drugs.
The aim of this study was to determine the antimicrobial resistance of isolates, isolated from material originating from poultry farms during the one-year period (2014) in the territory of Nis and South Moravian epizootiological area. For the isolation and identification of causers, the standard microbiological methods according to EN ISO 6579: 2008 Annex D were used. Antimicrobial resistance is tested on 72 isolates of Salmonella spp. Examination of antimicrobial resistance was carried out by the disk diffusion method on Mueller-Hinton agar and interpreted according to the CLSI standard. The paper used thirteen types of antibiotic discs (Bioanalyse): enrofloxacin, norfloxacin, florfenicol, ceftiofur, amoxicillin, colistin, erythromycin, gentamicin, neomycin, kanamycin, flumequine, lincomicin/spectinomicin and sulfamethoxazole/trimethoprim.
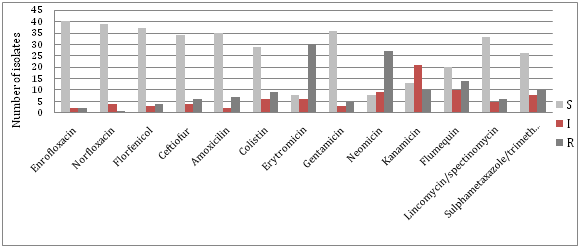
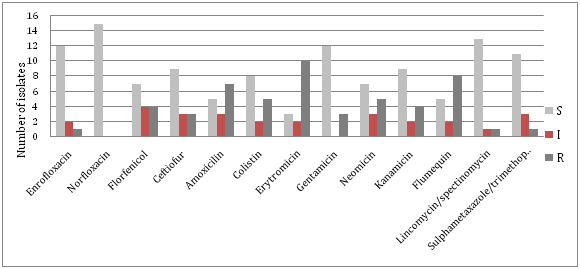
The results indicate the existence of a high percentage of resistance to some antimicrobial drugs. The greatest observed resistance was to erythromycin(68% at strain S. Enteritidis and 67% at S. Infantis), neomycin (61% at strain S. Enteritidis and 33% at S. Infantis), flumequine(32% at strain S. Enteritidis and 53% at S. Infantis), strains of S. Enteritidis showed a great resistance to kanamycin and sulfapreparations(23%) and colistin(20%), while the strains of S. Infantis were resistant in a large percentage to amoxicillin(47%) and colistin(33%). There is slightly less resistance to other antibacterial drugs, and only a few isolates were resistant to quinolones and gentamycin. The emergence of resistant strains of Salmonella can result in the possibility of treating animals and humans and therefore a more rigorous implementation of measures for the suppression of salmonellosis in animals is required.
Keywords: antibiotics, antimicrobial resistance, Salmonella spp., poultry
Salmonellosis is a zoonosis, an infectious disease of humans and animals caused by bacteria of the genus Salmonella. The genus Salmonella (S.) is divided into two species: Salmonella enterica and Salmonella bongori. More than 99% of known serotypes of Salmonella and all the most important serotypes that are pathogenic for humans belong to the species of Salmonella enterica. Certain serotypes are "host-specific", have a small host range and can cause serious systemic infections (S. Gallinarum in poultry, S. Typhi in humans or S. Abortus-ovis in sheep). The second group includes "host restricted" serotypes, they are adapted to a wide host range (S. Dublin causes serious disease of cattle, but it may also infect humans). There is a third group named "generalist", which includes the ubiquitous serotypes with a broad host range (S. Enteritidis, S. Typhimurium, S. Infantis, S. Hadar). Ubiquitous serotypes rarely cause systemic disease in animals and humans, but they are able to colonize the gastrointestinal tract of many species of hosts. Due to frequent colonization and fecal excretion of animals bred for human consumption, ubiquitous serotypes enter the food chain and can cause salmonellasis cases in humans.1
According to the World Health Organization in 1995 in 191 member countries, the three most common serotypes of Salmonella isolated from humans were S. Enteritidis, S. Typhimurium and S. Typhi (76.1% of all registered cases). In Europe, where there are no endemic infections with S. Typhi, the most common serotypes were S. Enteritidis (64.5%), S. Typhimurium, S. Infantis, S. Virchow and S. Newport.2 People can become infected with Salmonella by direct contact with animals or by the fecal-oral route. However, the most important and most common route of transmission of salmonellosis is through contaminated food of animal origin. Infection of humans by serotypes S. Enteritidis is frequently associated with the consumption of chicken eggs and poultry meat. In recent years, the worldwide decline in the number of cases of Salmonella infections was observed in comparison to the previous period, likely due to increased awareness and implementation of programs for the control of Salmonella on farms and in the production process,3 but according to some authors the drop in the number of patients is a temporary fluctuation.4
Salmonellosis of poultry is a particularly significant problem in public health. The mere presence of Salmonella in poultry flocks and the emergence of salmonellosis caused great economic losses in poultry production. Direct damage can occur at every stage of production. Infections of adult poultry often pass asymptomatically; however, at this stage production costs significantly increase due to measures taken to prevent the spread on the progeny and the people. While other serotypes of Salmonella can colonize the gastrointestinal organs of livestock, only S. Typhimurium and S. Enteritidis can colonize the reproductive organs and thus contaminate eggs in the course of formation. Additionally, S. Enteritidis is able to survive in the egg after spawning, can be found in the egg white and yolk, but is more common in the egg white.5
Given the widespread occurrence of Salmonella in nature, the customary treatment of animals is by antimicrobial agents. As a consequence resistance to different drugs develops. Resistance interferes with the treatment of people, but it also creates a cumulative environmental problem. During the contact of resistant salmonella and saprophytic bacteria in the gastrointestinal tract, the resistance genes from Salmonella can be transferred into the genome of saprophytic bacteria sensitive to antibiotics through horizontal plasmid transportation. In this way, saprophytic and/or opportunistic bacteria acquire point mutations in specific genes of the genome of its own and become resistant.6 Until recently it was thought that Salmonella are "antibiotic foreseeable" bacteria, that are always sensitive to all antibiotics and that do not develop resistance. The first cases of Salmonella resistant to antibiotics have emerged in the mid-eighties in strains of S. Enteritidis and S. Typhimurium. Initially the resistance existed only to ampicillin and tetracycline. In the mid-nineties of the last century appeared a multiresistant strain of S. Typhimurium (DT 104), which was resistant to ampicillin, fluorinated quinolones, streptomycin, sulfonamides, tetracyclines, trimethoprim, spectinomicin and disinfectants. This strain in the Scandinavian countries caused great economic losses due to mortality on swine farms, and spread on the other animal species and humans, causing high mortality.7,8
In recent years, the emergence and spread of antibiotic resistant strains of zoonotic bacteria pose a serious public health problem. Everything is the result of a long-term antibiotic therapy, which leads to the emergence of antimicrobial resistance to one or more antibiotics (multiresistance). It has been shown that the resistance genes are located on plasmids, that these genes are often grouped into integrons and that the transfer of resistance to progeny of bacteria is very common.9,10
In Serbia, in the ten-year period from 2004 to 2013 the declining trend of the incidence of antimicrobial resistance for S. Enteritidis (7%) and S. Hadar was observed. Testing isolates of S. Typhimurium registered the significant occurrence of resistance, especially multiresistance. It was found about 30% of resistant isolates with distinct heterogeneity of phenotypes and high incidence of multiresistant isolates (83%). Also, the resistance in the strain of S. Infantis11 is getting more common. The European Union's Council Directive 2003/99/EC prescribes mandatory monitoring of Salmonella resistance as well as other zoonotic organisms. Whereas the influence of pathogenic effects of Salmonella on human population is of great importance, the aim of this study was to determine the antimicrobial resistance of isolates, isolated from materials originating from poultry during the one-year period, in 2014, on the territory of Nis and South Moravian epizootiological areas.
As a material for testing, isolates of Salmonella spp. collected in 2014 from material originating from poultry farms (samples of faeces, carcasses of poultry, washers and samples from the environment) were used. Materials are reviewed in accordance with the Regulations on establishing measures for the early detection, diagnosis, prevention of spreading, suppression and eradication of infections of poultry by certain Salmonella serotypes12 and on the basis of contractual cooperation with the owners, in the course of ordinary and extraordinary control as well as during an inspection. In 2014 a total of 2113 samples were examined for the presence of Salmonella.
For the isolation and identification of causers, the standard microbiological methods according to EN ISO 6579:2008 Annex D were used. The 25g of samples each were poured with 225ml of BPW (Buffered Peptone Water, HiMedia Laboratories, India) and incubated for 18-22h at a temperature of 37°C. After incubation, 0.1ml was sieved on MSRV (Modified semi-solid Rappaport Vassiliadis, HiMedia Laboratories, India) agar and incubated for 24h at a temperature of 41.5°C, and 1ml of BPW is sieved in MKTTn (Muller-Kauffmann-Tetrathionate Broth Novobiocin-Muler Kaufman tetrathionate broth, Biokar Diagnostics, France) and incubated during 24h at a temperature of 37°C. After the expiry of incubation, the broth cultures were re-sieved on XLD (Xylose Lysine deoxycholate Agar, HiMedia Laboratories, India), and BGA (Brilliant Green Agar, HiMedia Laboratories, India), and incubated for 24h at a temperature of 37°C. Obtained isolates were purified and biochemically and serologically tested for Salmonella species affiliation. We identified 72 isolates of Salmonella spp. and all isolates were tested for antimicrobial resistance.
Investigation of antimicrobial resistance was carried out by the disk diffusion method on Mueller-Hinton agar (HiMedia Laboratories, India) and interpreted according to the CLSI standard - Clinical and Laboratory Standards Institute documents M100-S22.13 Thirteen types of antibiotic discs were used: enrofloxacin (ENR-5μg), norfloxacin (NOR-10μg), florfenicol (FFC-30μg), ceftiofur (FUR-30μg), amoxicillin (AX-25μg), colistin (CT-10μg), erythromycin (E-15μg), gentamycin (GN-10 μg), neomycin (N-30μg), kanamycin (K-30μg), flumequine (FLM-30μg), lincomycin /spectinomycin (LCS-109μg) and sulfamethoxazole/ trimethoprim (SXT-25μg ). For quality control, we used the reference cultures of Salmonella enteritidis ATCC® 13076TM.
In 2014, on a total of 2113 samples originating from poultry were examined (samples of faeces, carcasses of poultry, washers and samples from the environment), of which 72 (3.40%) were positive for the presence of Salmonella. Table 1 shows that the most frequently isolated Salmonella serotypes were S. Enteritidis 44 samples (61.11%), S. Infantis 15 samples (20.83%) and S. Typhimurium 7 samples (9.72%). Much less present were other serotypes of Salmonella - S. Montevideo, S. Newport and S. Paratyphi A.
|
S |
I |
R |
Enrofloxacin |
40 |
2 |
2 |
Norfloxacin |
39 |
4 |
1 |
Florfenicol |
37 |
3 |
4 |
Ceftiofur |
34 |
4 |
6 |
Amoxicilin |
35 |
2 |
7 |
Colistin |
29 |
6 |
9 |
Erytromicin |
8 |
6 |
30 |
Gentamicin |
36 |
3 |
5 |
Neomicin |
8 |
9 |
27 |
Kanamicin |
13 |
21 |
10 |
Flumequin |
20 |
10 |
14 |
Lincomycin/Spectinomycin |
33 |
5 |
6 |
Sulphametaxazole/Trimethoprim |
26 |
8 |
10 |
Table 1 Resistance in S. Enteritidis strains to antibiotics compared to the total number of examined isolates S. enteritidis
By testing sensitivity of isolated strains of S. enteritidis to antimicrobial drugs, resistance to fluorinated quinolones was less than 5%, or only two strains showed resistance to enrofloxacin and norfloxacin. The high prevalence of resistance is found to erythromycin (68%), neomycin (61%), flumequine (32%), kanamycin (23%), sulfamethoxazole / trimethoprim (23%) and colistin (20%). Other results related to resistance to antibiotics in strains of S. Enteritidis in comparison to the total number of examined isolates of S. enteritidis are shown in Figure 1.
S. infantis shows good sensitivity to fluorinated quinolones, cephalosporins, lincomycin / spectinomycin and sulfa preparations, while the great resistance occurs to erythromycin (67%), flumequin (53%), amoxicillin (47%), neomycin and colistin (by 33%) (Table 2). The results of presence of antibiotic resistance in strains of S. Infantis are shown in Figure 2.
|
S |
I |
R |
Enrofloxacin |
12 |
2 |
1 |
Norfloxacin |
15 |
0 |
0 |
Florfenicol |
7 |
4 |
4 |
Ceftiofur |
9 |
3 |
3 |
Amoxicilin |
5 |
3 |
7 |
Colistin |
8 |
2 |
5 |
Erytromicin |
3 |
2 |
10 |
Gentamicin |
12 |
0 |
3 |
Neomicin |
7 |
3 |
5 |
Kanamicin |
9 |
2 |
4 |
Flumequin |
5 |
2 |
8 |
Lincomycin/Spectinomycin |
13 |
1 |
1 |
Sulphametaxazole/Trimethoprim |
11 |
3 |
1 |
Table 2 Resistance in S. Infantisstrains to antibiotics compared to the total number of examined isolates S. Infantis
The number of salmonella isolated from the strain of S. Typhimurium was small to assess antimicrobial resistance. By testing sensitivity of isolated strains of S. Typhimurium to antibacterial drugs, a complete sensitivity to fluorinated quinolones and gentamicin and small resistance of aminoglycoside antibiotics, lincomycin/ spectinomycin and sulfa preparations were found. The highest rates of resistance were found in erythromycin, flumequine, colistin and cephalosporins.
The study of resistance to antimicrobial agents in two isolates of S. Newport, and in three isolates of S. Montevideo showed resistance only to amoxicillin and flumequine. Salmonella Paratyphi A is only to be resistant to amoxicillin. When we compare our data with data from the literature, it can be seen that in other regions of our country and in other countries S. Enteritidis is most frequently isolated Salmonella in both animals and humans.11,14,15 Salmonella Infantis is in recent years increasingly isolated in animals and in human medicine.11,16
The most frequently isolated strain in poultry flocks, S. Enteritidis, showed considerable resistance to some antibiotics. It is the result of a long-term use of certain antibiotics in poultry. For S. Enteritidis there is great resistance to the following antibiotics: erythromycin, neomycin, flumequine, kanamycin and colistin. If we compare our results with the results of other authors in the past, we can conclude that resistance among our isolates is much higher especially to erythromycin, flumequin, colistin and sulfa preparations.17,18 Resistance to fluorinated quinolones is small despite the widespread use in poultry, particularly resistance to enrofloxacin. Enrofloxacin is not an effective drug in the treatment of salmonellosis of birds because after stopping the therapy salmonella colonize the intestinal tract again. This medicine was withdrawn from use in poultry in the United States because its use can develop resistance in Campylobacter species.8 For the emergence of resistance to quinolones in S. Enteritidis, gene mutation girA is responsible. The same mutation leads to the emergence of multiresistant strains. According to research conducted in our country multiresistant Salmonella strains are present in poultry, and the findings of some authors point to the need for the introduction of the laboratory practice of adequate system of molecular standardization of strains and monitoring of resistance, particularly resistance to quinolones in Salmonella.19,20
Monitoring of resistance in S. Infantis is very important due to the frequent occurrence of this serotype that is mainly derived from poultry. According to research Velhner et al.21 PFGE analysis showed that in Serbia S. Infantis isolates have similar genetic profile indicating a clonal spread of resistance.21
The problem with resistance in the strain of S. Typhimurium is serious because of the emergence of multiresistant strains of S. Typhimurium DT104, DT204, DT193 etc. which give rise to serious alimentary toxicoinfections and large losses in livestock.8,18
The growing phenomenon of resistance to antimicrobial medicines and particularly reduced sensitivity to cephalosporins and fluoroquinolones narrows drugs choice for effective antibiotic therapy and leads to problems with the treatment of severe forms of salmonellosis in humans. Resistance not only interferes with the treatment of humans and animals, but also creates a cumulative environmental problem. In the control of salmonellosis, what is of great importance is risk assessment and monitoring of different sources of infection and ways of its dissemination, system control, good hygiene practices in animal breeding and food production as well as vaccination of animals are necessary. There is a real need for a global initiative and establishing control over potentially dangerous bacterial strains and more rigorous implementation of measures to combat them in animals. Control measures include disinfection of buildings and equipment, rodent control, vaccination, treatment, and laboratory testing. Based on the research of some authors [19,20], there is a need for the application of molecular methods in antibiotic resistant strains in order to reveal the mechanisms of resistance occurrence and origin of strains (epizootiological and possibly epidemiological) or the origin of their resistant phenotypic traits.
Based on our results we can conclude that salmonella strains isolated from samples originating from poultry show the existence of resistance to certain antimicrobial drugs. By testing sensitivity of isolated S. Enteritidis strains to a panel of thirteen antibiotics, a high frequency of resistant isolates and multiply resistant isolates was established. The most common are resistances to erythromycin (68%), neomycin (61%), flumequine (32%), sulfapreparations, kanamycin (23%) and colistin (20%). Similar results are found with isolates S. Infantis, the largest resistance was observed to erythromycin (67%), flumequine (53%), amoxicillin (47%), neomycin and colistin (33%). For other antibacterial drugs there is much less resistance. Resistance of isolated Salmonella from our region to fluoroquinolones is low (less than 5%) which is good as the fluoroquinolones are often used in human therapy.
None.
Author declares that there is no conflict of interest.

©2016 Manic, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.